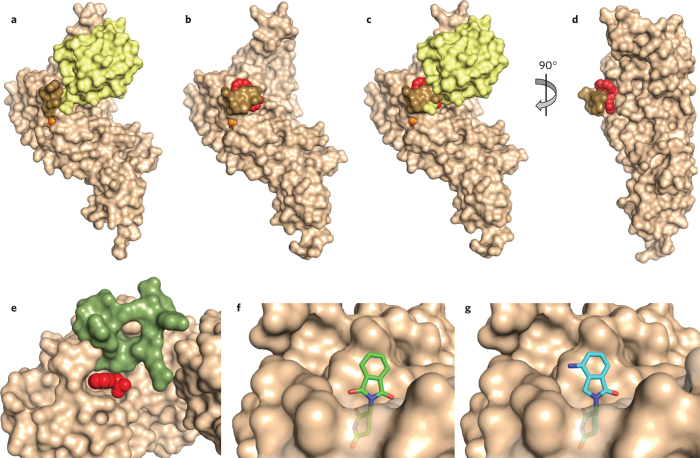
Figure 3. Structural basis of DUB and E3 modulation by small molecules.
(a,b) Structure of ubiquitin aldehyde (a; yellow) or GRL0617 (b; red) bound to the deubiquitinating enzyme PLpro (tan). Images derived from PDB code 4MM3 (ref. 25) and 3E9S (ref. 9), respectively. The active site cysteine is shown in orange, and a loop that closes over the inhibitor is shown in brown. Notice how this loop changes position to close over the inhibitor. (c) Ubiquitin aldehyde is overlaid with the image in b to demonstrate that the loop clashes with the position of ubiquitin when it closes over the inhibitor. (d) View from b is rotated to better show position of inhibitor binding in the site of loop closure. (e) Auxin (red) binding to TIR1 ubiquitin ligase (tan) completes the binding site for the substrate (green); derived from PDB code 2P1Q (ref. 13). Some residues of TIR1 have been removed to better visualize the auxin binding site. (f) Binding of thalidomide (green) to the E3 cereblon (tan) is believed to block binding of substrates at the same site. (g) Binding of lenalidomide (cyan) to cereblon (tan) is believed to promote binding of new substrates owing to its difference in chemical structure relative to thalidomide, shown projecting from the surface of cereblon. Structures in f and g are derived from PDB codes 4CI1 and 4CI2 (ref. 18), respectively.