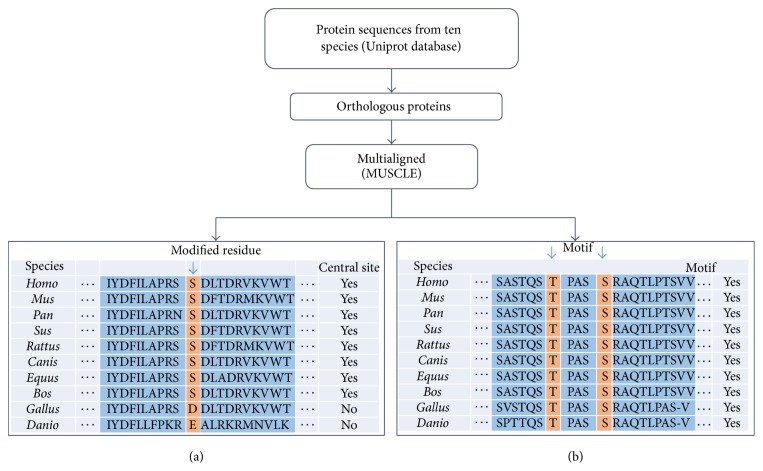
Figure 1.
The workflow to calculate the evolutionary conservation score of modified sites and motifs. Protein sequences of ten species were downloaded from Uniprot database and then each group of orthologous proteins was multialigned by MUSCLE software. (a) Conservation scores of modified sites were calculated. (b) After extracting overrepresented motifs, proteins containing these motifs were selected and conservation of motifs in these proteins was calculated.