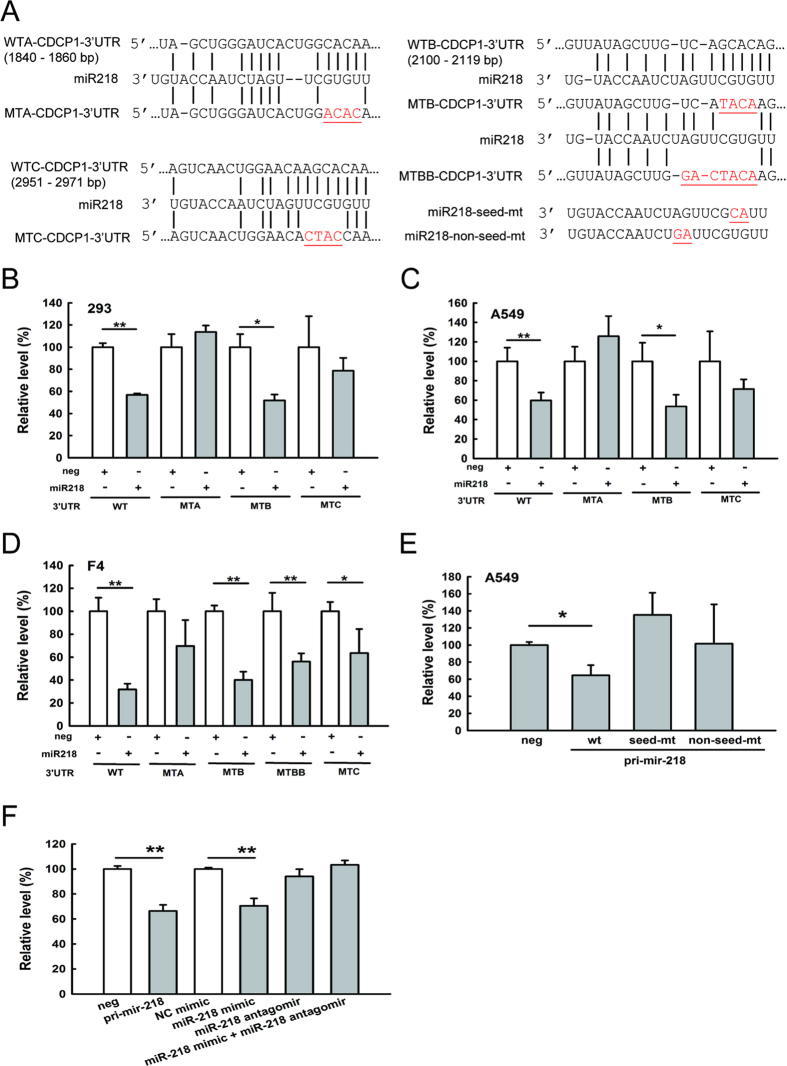
Figure 3. CDCP1 is a target of miR-218.
(A) Schematic representation of miR-218 targeting the CDCP1 3′-UTR. Three potential miR-218 binding sites are located at 1,840–1,860 bp (site WTA), 2,100–2,119 bp (site WTB), and 2,951–2,971 bp (site WTC) from the transcription start site of CDCP1. Mutation sequences at these 3 miR-218 binding sites and miR-218 mutation at seed or non-seed sites are marked with red letters and underlines. (B–D) Luciferase assays of miR-218 binding to the wild-type and mutated CDCP1 3′-UTR in HEK 293 cells (B), A549 cells (C), and F4 cells (D). Cells were co-transfected with the plasmid of pri-mir-218 as miR-218, the firefly luciferase construct of CDCP1 3′-UTR, and the Renilla luciferase control for the dual-luciferase assay. The relative luciferase activity represents the dual luciferase activity ratio (firefly/Renilla luciferase). WT: wild-type; MTA, MTB, MTC: mutation at site A, B, or C, respectively; MTBB: extended length of mutant sequence at site B. (E) Luciferase assays of A549 cells co-transfected with pri-mir-218 wild-type or mutants, the firefly luciferase construct of CDCP1 3′-UTR, and the Renilla luciferase control. (F) Luciferase assays of miR-218 binding to the wild-type CDCP1 3′-UTR in A549 cells. Pri-mir-218, miR-218 mimic, or miR-218 antagomir were included in the assays. *P < 0.05. **P < 0.01.