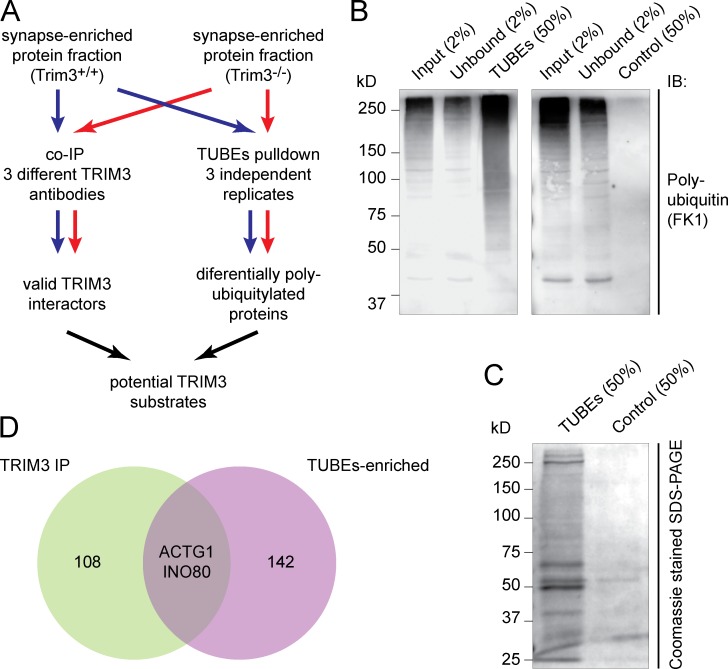
Figure 4.
Identification of potential TRIM3 substrate proteins. (A) Strategy for identifying TRIM3 substrates. Hippocampal synapse-enriched protein fractions were used as input for both immunoprecipitation (left) and polyubiquitin affinity pull-down (right). Samples from Trim3−/− mice served as controls. All samples were analyzed by quantitative mass spectrometry and potential TRIM3 substrates were identified by ranking and comparing results from all six samples. (B) Validation of the TUBEs pull-down approach. Input, unbound, TUBEs pull-down (50%) and control pull-down (agarose beads only) fractions were resolved on SDS-PAGE and immunoblotted for polyubiquitin. A strong enrichment for polyubiquitylated proteins (50–250 kD) was observed in the TUBEs pull-down fraction (left), but not in the control pull-down fraction (right). (C) TUBEs pull-down samples were highly enriched in proteins compared with control pull-down samples. (D) Venn diagram indicating the proteins that were identified as potential TRIM3 substrates. Coimmunoprecipitation identified 110 TRIM3-interacting proteins, whereas TUBEs pull-down identified 144 proteins that were more polyubiquitylated in wild-type mice compered to Trim3−/− mice (Table S2). Only two proteins were identified in both experiments: γ-actin (ACTG1) and DNA helicase INO80.