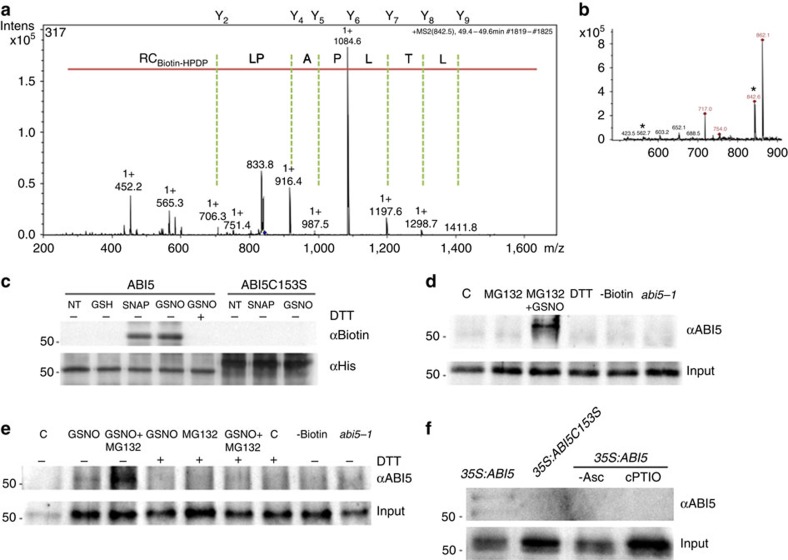
Figure 3. S-nitrosylation of ABI5 in vivo and in vitro.
(a) Mass spectrometric analyses identify C153 as the S-nitrosylation site. MS/MS spectra of C153 from the tryptic fragment QGSLTLPAPLCR (peptide MS/MS spectra shown with Cys modified by biotin-HPDP). (b) The LC–MS spectra of the corresponding peaks (*562 m/z (3+) and 842,49 m/z (+2)) of this peptide fragment is shown in the inset. (c) The C153S mutation blocks S-nitrosylation of ABI5. In vitro S-nitrosylation of wild-type ABI5 and mutant ABI5C153S recombinant proteins by the NO donors GSNO (200 μM) and SNAP (200 μM). This modification is reversed by treatment with DTT (20 mM). No signal was observed with glutathione (200 μM) treatment showing specificity of the biotin-switch assay. ABI5 protein loading was detected by anti-His antibody. (d,e) S-nitrosylation of ABI5 induced by GSNO in after-ripened seed extracts. Samples were initially immunopurified with anti-biotin before immunoblot analysis of ABI5 protein levels in seed extracts of Col-0 (d), 35S:ABI5 (e) and abi5-1 untreated (C) or treated with the indicated compounds. No signal was observed in the absence of biotin (−Biotin) or after DTT (20 mM) treatment. Actin protein levels are shown as a loading control. (f) In-vivo S-nitrosylation of ABI5 in abi5-1;35S:cMyc-ABI5 and abi5-1;35S:cMyc-ABI5C153S after-ripened seed extracts 24 h after proteasome inhibitor MG132 (100 μM) incubation. Immunoblot analysis of in vivo ABI5 protein levels after immunopurification of S-nitrosylated proteins. No signal was observed in the absence of sodium ascorbate (−Asc) or after cPTIO (1 mM) treatment. Input protein levels were also determined using anti-ABI5 anti-serum.