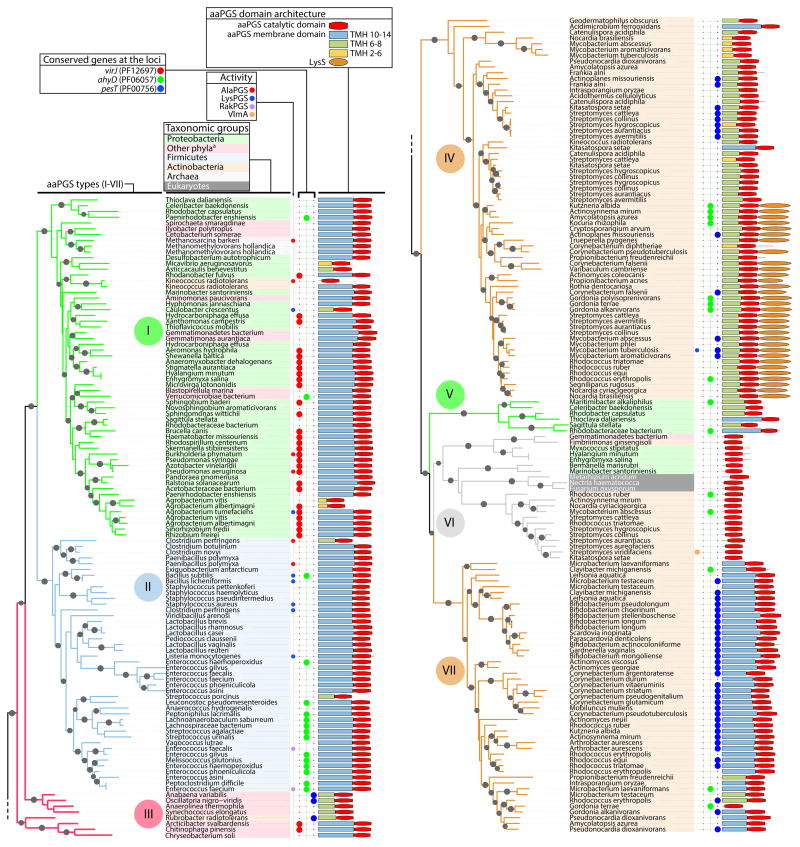
Fig. 1. Phylogenetic analysis of aaPGS homologs.
Phylogenetic tree was constructed based on alignment of 160 residues in the C-terminal domain of 253 aaPGS homolog sequences using the program FastTree. Organism names are shown on a color background delineating the major taxonomic groups. The functions of characterized homologs (LysPGS, AlaPGS, RakPGS, VlmA), occurrence of adjacently encoded hydrolytic enzymes (VirJ, AhyD, PesT), and aaPGS domain architectures are indicated for each organism. When present, the number of predicted transmembrane helices (TMH 2–14) in the membrane domain is color-coded. Sequence identification (GI) numbers are shown in Fig. S3. aBacteria from these phyla belong to one of the following: Fusobacteria, Gemmatimonadetes, Planctomycetes, Bacteroidetes, Chloroflexi, Cyanobacteria, Spirochaetes, Synergistetes, Verrucomicrobia.