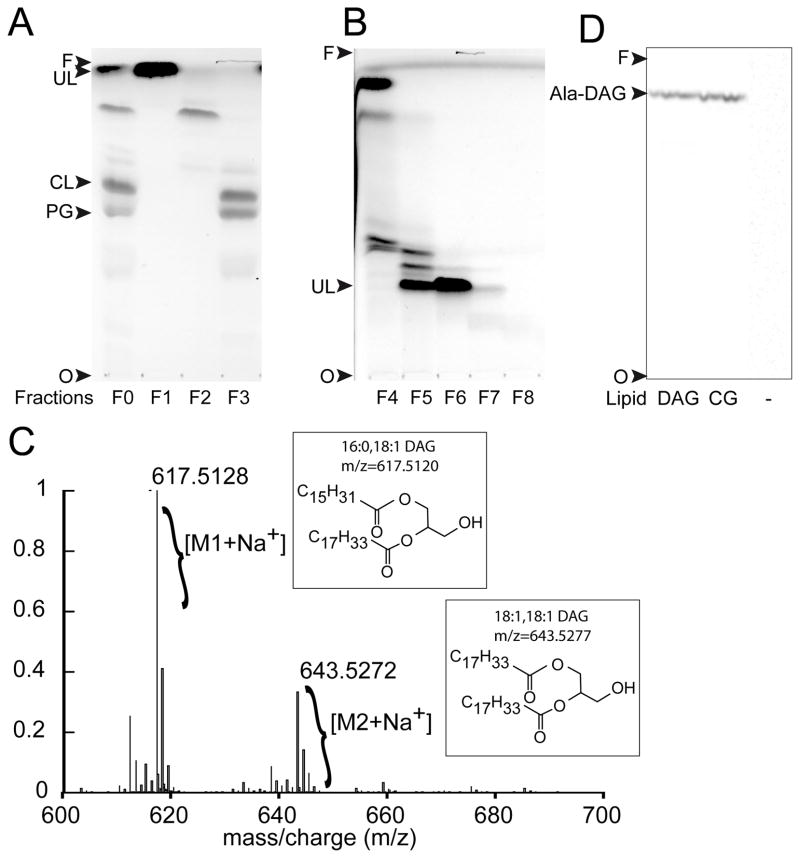
Fig. 3. Identification of DAG as a lipid substrate of Cg1103.
(A) TLC analysis (using solvent A for the mobile phase) of lipids extracted from the wild-type strain of C. glutamicum, before (F0) and after (F1–F3) fractionation of lipid classes by flash chromatography. TLC spots were isolated and tested for tRNA-dependent lipid alanylation activity to identify the unknown lipid (UL) substrate of Cg1103. (B) Lipids in fraction F1 were further separated using a second step of flash chromatography on silica gel. Resulting fractions (F4–F8) were analyzed by TLC using solvent B for the mobile phase. (C) ESI/TOF mass spectrum of purified UL from fraction F6. (D) tRNA-dependent lipid alanylation activity of Cg1103483–832 was assayed in the presence of commercial 18:1-16:0 DAG, lipids extracted from C. glutamicum (CG), or in the absence of lipids (−). Lipids modified with [14C]Ala were analyzed by TLC (in solvent A), and visualized by phosphorimaging. Ala-DAG: alanyl-diacylglycerol, CL: cardiolipin, PG: phosphatidylglycerol, O: TLC origin, F: solvent front.