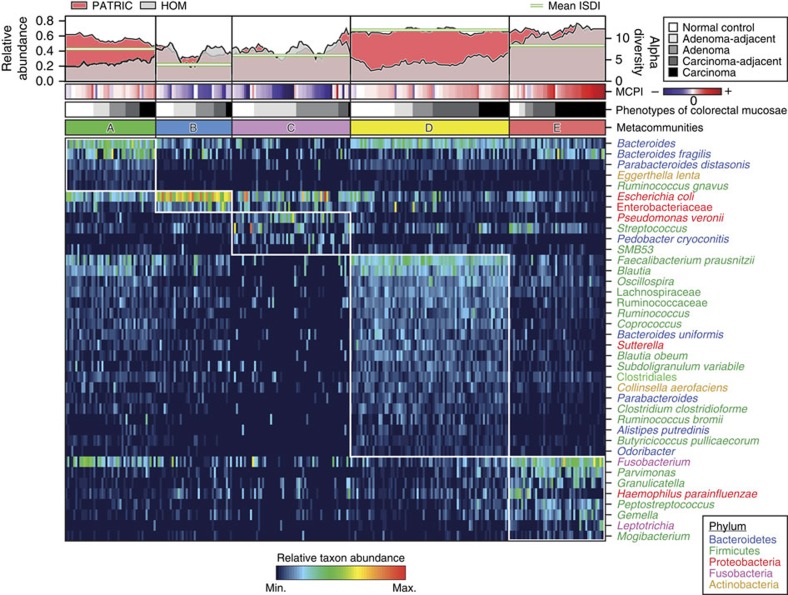
Figure 1. Characterization of 16S rRNA gene catalogue for mucosal microbial communities in colorectal carcinogenesis.
Fitting microbiome data to DMM models defined five metacommunities. Reads that are considered as being potentially originated from oral strains or known pathogenic strains in the human gut were classified against the 16S rRNA gene collections from the Human Oral Microbiome (HOM; version 13) database and PATRIC bacterial pathogen database as defined by pseudo-bootstrapped (n=1,000) confidence scores of 100 at species-level taxa or deeper, using the naive Bayesian classifier. The panels of metacommunity markers are ranked in the descending order of linear discriminant analysis scores from top to bottom. Columns represent microbiome profiles (arcsine square root-transformed) of 269 mucosal biopsies from individuals with or without adenoma or adenocarcinomas. (MCPI<0 for changes characteristic of adenomas; MCPI>0 for changes characteristic of carcinomas).