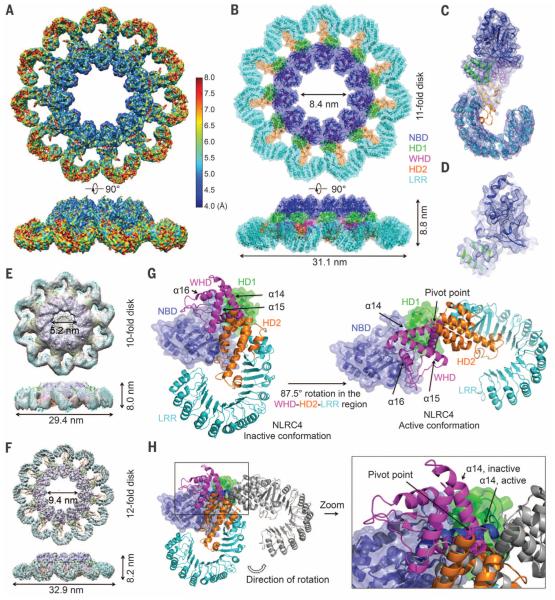
Fig. 2. Cryo-EM structure determination and conformational activation of NLRC4.
(A) Cryo-EM map of the C11 PrgJ-NAIP2-NLRC4Δ complex colored with local resolution calculated by ResMap using two separately refined half maps. (B) Superimposed ribbon diagram and transparent surface of the C11 NLRC4Δ structure. (C) Cryo-EM density superimposed with one NLRC4Δ subunit. (D) A close-up view of the structure of the NBD of NLRC4Δ superimposed with the cryo-EM density. (E and F) Cryo-EM maps and fitted NLRC4Δ models for the C10 reconstruction at 12.5 Å resolution (E) and the C12 reconstruction at 7.5 Å resolution (F). (G) The WHD-HD2-LRR domain of NLRC4 swings 87.5° to transit from the inactive conformation (left, PDB ID 4KXF) to the active conformation (right). NBD and HD1 are shown in superimposed ribbon diagram and transparent surface, and the WHD-HD2-LRR module is shown in ribbon diagram. (H) Superimposed inactive (colored) and active (gray, except for a14 helix, which is in dark blue) conformations of NLRC4Δ. The α14 helices in the two conformations are labeled to show the relative rotations and the rotational pivot point.