Fig. 1. Characterization of (1-3,1-4)-β-glucan structure in Nicotiana benthamiana leaves.
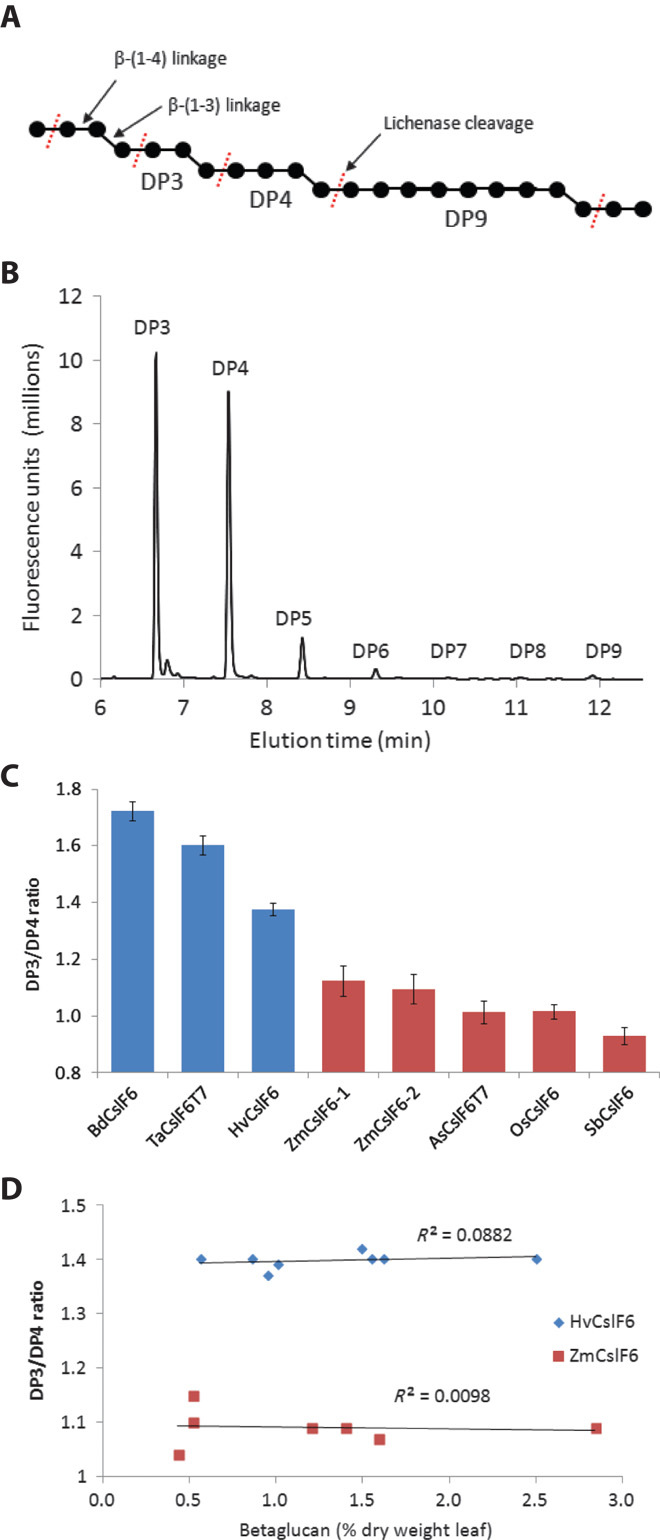
(A) Structure of (1-3,1-4)-β-glucan. Glucose molecules are represented by black dots with β1-4 linkages (straight lines) or β1-3 linkages (angled lines). The bacterial enzyme lichenase specifically cleaves (1-3,1-4)-β-glucan at a β1-4 linkage after a β1-3 linkage as indicated by dashed red lines, producing short oligosaccharides with a DP from DP3 to DP9. (B) Oligosaccharides released by lichenase digestion of betaglucan were fluorescently labeled with APTS and separated by capillary electrophoresis. (C) The source of the CslF6 gene affects the DP3/DP4 ratio of the (1-3,1-4)-β-glucan produced in N. benthamiana leaves. The CslF6 proteins can be classified into two groups: those that produce a (1-3,1-4)-β-glucan with a high (>1.3) DP3/DP4 ratio (Bd, Brachypodum distachyon; Ta, Triticum aestivum; Hv, Hordeum vulgare; shown in blue) or a low (<1.1) DP3/DP4 ratio (Zm, Zea mays; As, Avena sativum; Os, Oryza sativa; Sb, Sorghum bicolor; shown in red). T7 is an 11–amino acid epitope tag at the N terminus of some of the CslF6 proteins—it has no effect on the amount or structure of the (1-3,1-4)-β-glucan. Results are averages ± SD from replicate measurements of between 2 and 11 independent experiments with each gene. (D) The DP3/DP4 ratio of the (1-3,1-4)-β-glucan produced by the HvCslF6 or ZmCslF6 proteins is very stable over a wide range of (1-3,1-4)-β-glucan concentrations in independent experiments. There is no correlation between the DP3/DP4 ratio and the amount of (1-3,1-4)-β-glucan produced.
