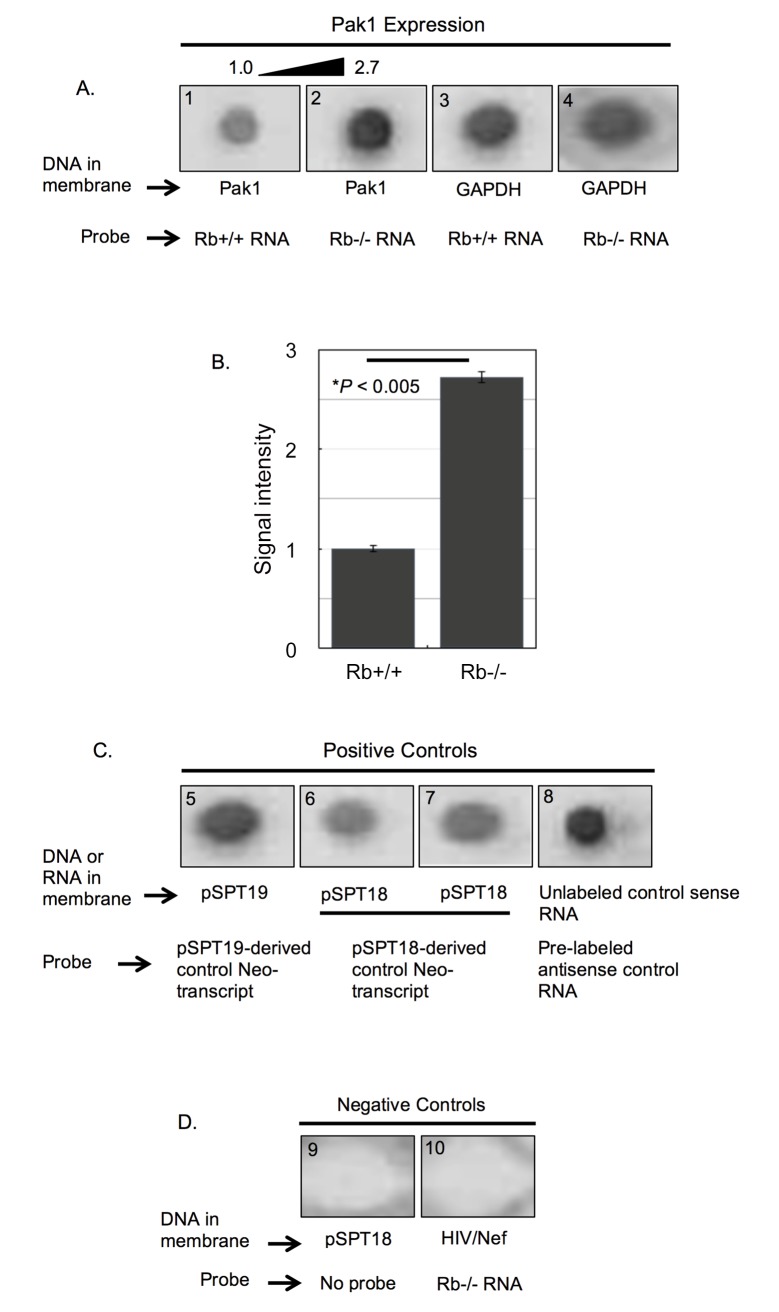
Fig 2. Rb represses the transcription rate of the Pak1 gene.
(A) Representative nuclear run-on assay from a triplicate experiment showing hybridization of Rb+/+ (blot 1) and Rb-/- (blot 2) DIG-labeled mRNA probes to a Pak1 cDNA immobilized in a membrane. These two probes were also hybridized to GAPDH cDNAs (blots 3 and 4). Hybridization signal intensity was stronger with the Rb-/- probe, indicating higher transcriptional rate of the Pak1 gene in Rb-/- nuclei. (B) To determine the differences in signal intensity, the chemioluminiscence in the membranes was quantified and analyzed using BioRad’s Quantity 1 Imaging Software and normalized against the signal intensities obtained when hybridizing to the GAPDH cDNAs (blots 3 and 4 in A). This quantitative analysis showed a statistically significant 2.7-fold increase in the signal intensity of the Rb-/- hybridization over the Rb+/+ hybridization. Each bar represents the mean of at least 3 independent experiments (±SE of the mean), with P < 0.005. (C) Strong signal intensities were observed in all positive controls in which pSPT19- and pSPT18-derived DIG-labeled control transcripts were hybridized to the parental plasmid from which they were transcribed (blots 5–7), as well as a pre-labeled antisense control RNA hybridized to its corresponding unlabeled sense RNA (blot 8). (D) As negative controls, either the probe was omitted from the hybridization reaction (blot 9) or a DIG-labeled mRNA probe from osteoblasts was hybridized to an irrelevant plasmid encoding the lentiviral protein Nef (blot 10).