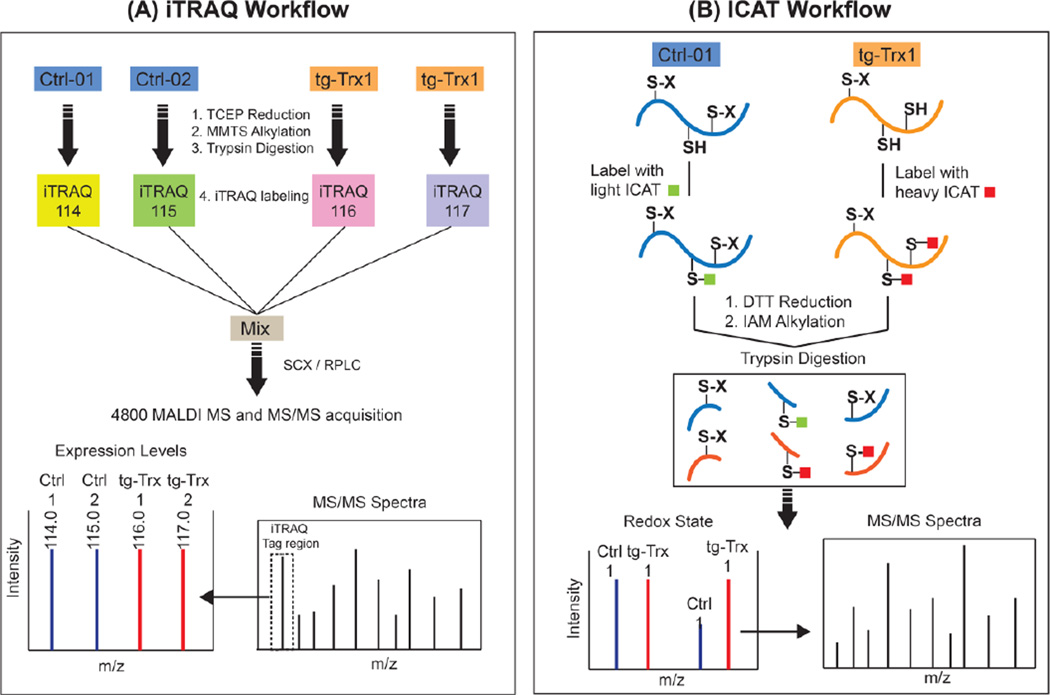
Figure 1. Typical workflow for ICAT and iTRAQ quantitation.
A) In the iTRAQ workflow, protein samples are first subjected to TCEP reduction, MMTS alkylation and trypsin digestion. The resulting tryptic peptides are then labeled by designated iTRAQ reagents separately. After quenching the reaction, labeled peptides are mixed and separated by multidimensional chromatography. Finally, MS data is acquired on a 4800 MALDI TOF/TOF in a data dependent acquisition mode. B) In the ICAT workflow, protein thiols are first labeled by either the light ICAT (control) or heavy ICAT (Trx1-overexpressing tissue) reagents. Protein disulfide bonds are then reduced by DTT and alkylated with IAM, sequentially. The labeled proteins are mixed, digested with trypsin and separated sequentially using SCX, avidin affinity and RPLC separations. ICAT-labeled peptides are identified and quantified by a 4800 MALDI-TOF/TOF mass spectrometer. Peptides containing Trx1-reduced cysteines had an ICAT H/L ratio larger than one, and can be quantified by the precursor peak intensity and identified by the MS/MS spectrum (Modified from Molecular & Cellular Proteomics, 2009 (8), 1674–1687 with permission).