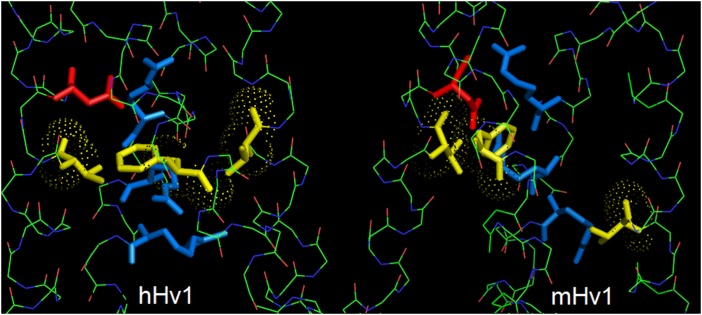
Fig. 2.
The hydrophobic gasket (three yellow amino acids) is aligned in the new hHV1 model based on the CiVSP crystal structure (Left), but not in the chimeric mHV1 crystal structure (Right). In each closed channel viewed from the side, the three S4 Arg are blue and the Asp in S1 that produces proton selectivity is red. The apparent misalignment in mHV1 could reflect a disturbance due to the spliced-in S2–S3 segment from CiVSP, which ends two positions below V174, the anomalous residue in mHV1. Alternatively, a different amino acid might complete the gasket in mHV1.