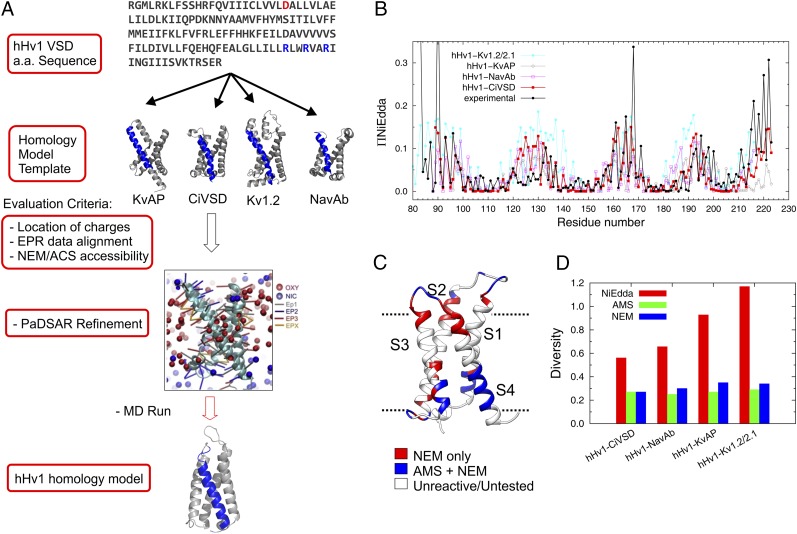
Fig. 3.
Structural modeling for hHv1-VSD at resting state. (A) Strategy of structural modeling by MD simulation. Starting from the sequence alignments, four structural models of hHv1-VSD were built on the templates of crystal structures of KvAP, CiVSD-WT, Kv1.2_chimera, and NavAb respectively. Each of the structural models was equilibrated with explicit lipid POPC and stabilized for 300 ns by MD simulation. The four MD stable models were evaluated with experimental data primarily with ΠNiEdda and NEM/AMS accessibility data. The structural model correlating best with experimental accessibilities was subjected to an additional MD simulation with constraints from present EPR spectroscopic data using an established computational method, PaDSAR (36). The constrained model was equilibrated in explicit lipid POPC for 400 ns, which serves our working model for the resting state of hHv1-VSD. (B) Predicted solvent accessibility from four structural models and the experimental NiEdda accessibility. (C) NEM/AMS accessibility data were mapped onto hHv1-VSD structural model from CiVSD-WT. (D) The correlation between predicted accessibilities and experimental accessibilities for each of the four hHv1-VSD structural models was evaluated by Kullback–Leibler divergence. The structural model from CiVSD-WT with lowest divergence was chosen to further construct the structural model for hHv1-VSD.