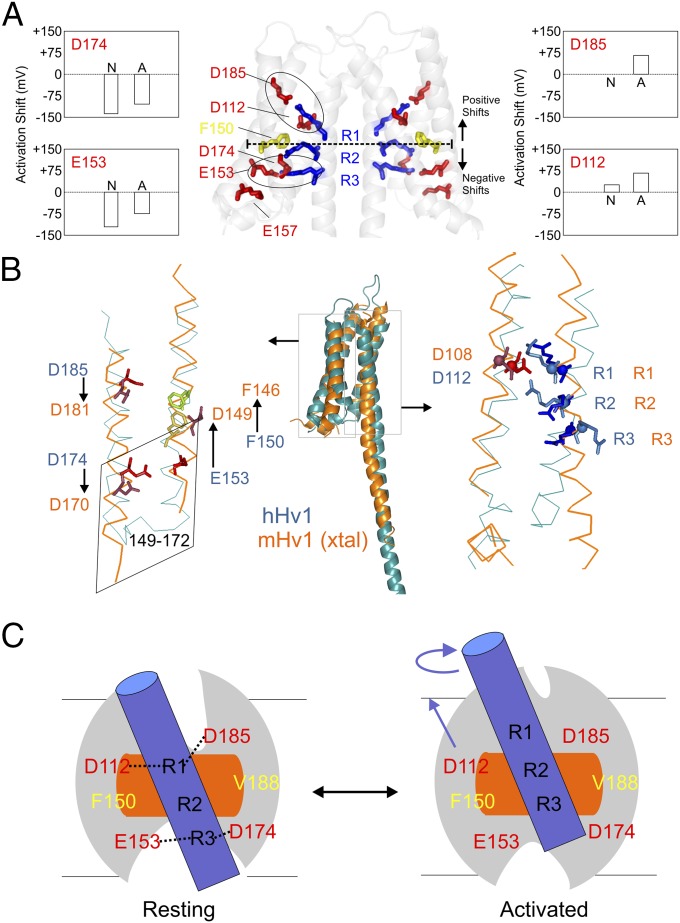
Fig. 6.
Model comparison and mechanistic implications. (A) Functionally critical residues in the hHv1-VSD dimer model. D174 and E153 are located below F150, the putative hydrophobic gasket, and D185 and D112 are located above. Neutralization of D174 and E153 causes left-shift in the G–V curve, whereas neutralization of D185 and D112 causes right-shift. The two electrostatic clusters D185-R1-D112 and E153-R3-D174 are highlighted with black circles. (B) Structural model of hHv1 was aligned with crystal structure of mHv-chimeric by the Cα of S1 + S2 + S3a. The residues on S1 (D112) and S4 (R1, R2, and R3) agreed very well (Right) with obvious differences of the residues on S2 and S3 between two models. To match mHv1-chimeric structure, the residues on S2 (D153 and F150) need to upshift one turn of helix, whereas the residues on S3 (D174 and D185) need to downshift one turn of helix. The chimeric region from CiVSD on mHv-chimera spanned from residues 149 to 172 (corresponding to residue 153–176 in hHv1), which are highlighted in the rhomboidal box. The structures of the identical sequence in CiVSD and mHv-chimera are distinctly different. (C) Proposed gating model of hHv1 according to the one-click gating mechanism in CiVSD. At resting-state hHv1-VSD in lipid bilayer, solvent penetrates from both intracellular and extracellular crevices leaving a focused electric field around R1 and R2 region at the level of hydrophobic gasket. hHv1 scaffold is shown by the gray oval; S4 by blue cylinder; hydrophobic gasket by orange rectangle. The two salt-bridge clusters (D112-R1-D183 and E153-R3-D174) are connected by black dotted lines. Up activation: the S4 of hHv1 rotates and moves three residues up.