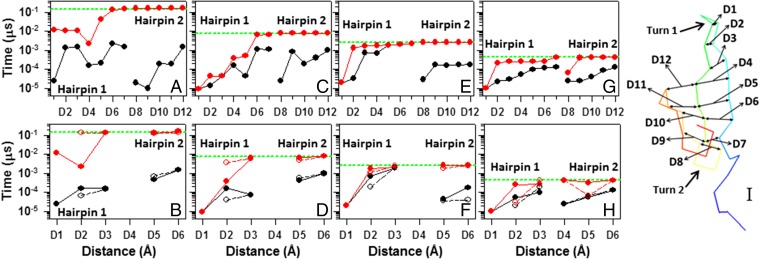
Fig. 2.
The first contact time (black circles connected by black lines) and stabilization time (red circles connected by red lines) vs. the distances between Cαs of selected pairs of residues of hairpin 1 (D1 → Ala14 and Gly16, D2 → Thr13 and Lys17, D3 → Lys12 and Thr18, D4 → Tyr11 and Tyr19, D5 → Glu10 and Tyr20, D6 → Thr9 and Tyr21, and D7 → Trp8 and Asn22) and hairpin 2 (D8 → Asn23 and Asp26, D9 → Asn22 and Glu27, D10 → Tyr21 and Ser28, D11 → Tyr20 and Thr29, and D12 → Tyr19 and Trp30) and also, vs. the distances between Cαs (Cβs are represented by white circles connected by dashed lines) of only nonpolar residues of hairpin 1 (D1 → Ala14 and Gly16, D2 → Tyr11 and Tyr19, and D3 → Trp8 and Tyr20) and hairpin 2 (D5 → Tyr20 and Pro33 and D6 → Tyr19 and Trp30) for (A and B) three-state, (C and D) two-state, and (E and F) downhill folding trajectories of the L26D mutant and (G and H) the downhill folding trajectory of the L26W mutant (D8 → Asn23 and Trp26 in G and D4 → Tyr21 and Trp26 in H). Structure of L26D is illustrated in I. Horizontal green dashed lines indicate the folding time of L26D and L26W.