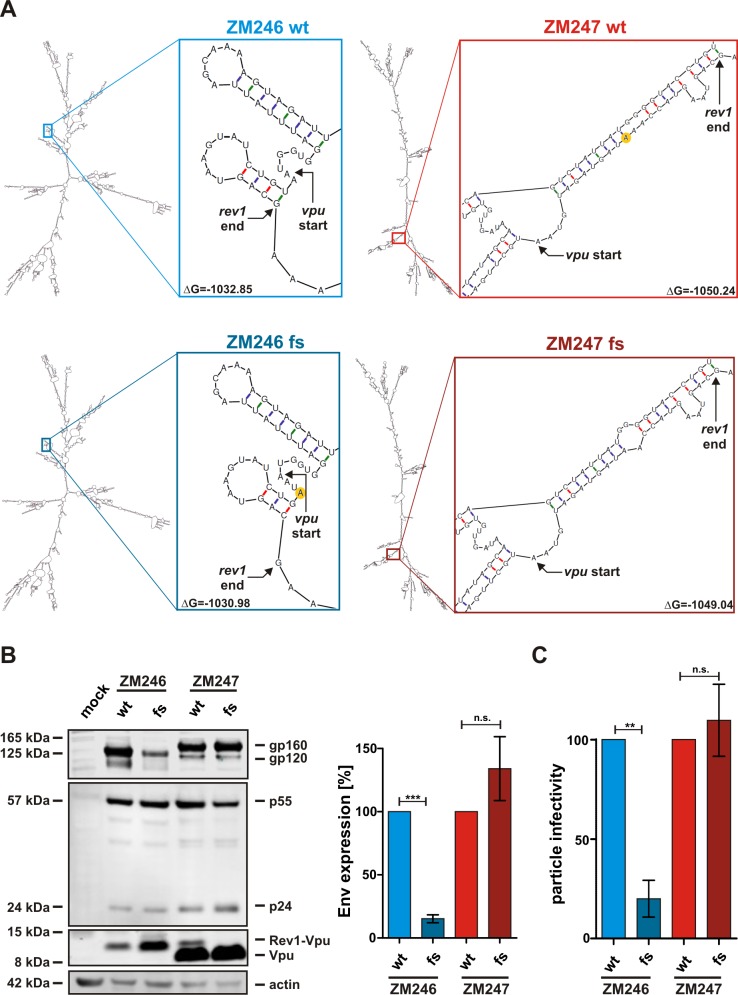
Fig 7. Effect of rev1-vpu frameshift mutations on mRNA structure and Env expression.
(A) mRNA structures of ZM246 wt, ZM246 fs, ZM247 wt and ZM247 fs were predicted using the Mfold web server for nucleic acid folding and hybridization prediction [37]. The putative mRNA structures of full length rev/vpu/env enconding mRNAs (using splice sites D1 and A4c) are shown on the left of each panel. Close-ups of the rev1-vpu intergenic region and the minimum free energy ΔG are shown on the right. The nucleotides that are deleted in ZM247 wt and inserted in ZM246 fs are highlighted in yellow. (B) Western blot analysis of HEK293T cells co-transfected with the proviral clones described in Fig 1A. gp160 and gp120 were detected using an antiserum raised against HIV-1 M subtype C 96ZM651. A representative Western blot is shown on the left. Total Env expression (gp120+gp160) was normalized to total Gag expression (p55+p24). The mean values (+/- SEM) of four independent transfections are shown on the right. (C) Particle infectivity was determined by infecting TZM-bl reporter cells with equal amounts of p24. Three days post infection, β-galactosidase activity was measured. The mean values (+/- SEM) of triplicate infections from two to four independent transfections are shown (***p<0.001; **p<0.01; n.s. not significant).