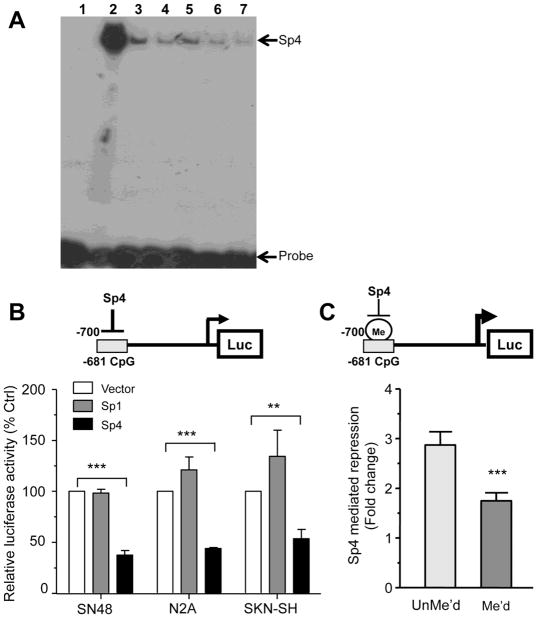
Figure 5. Sp4 repressor activity is decreased following methylation of 5-HT1A -681 CpG Site.
(A) Sp4 binds the Sp1-like element at CpG site -681 in vitro in EMSA assays. Bacterially-expressed Sp4 was incubated with a radio-labelled 5-HT1A CpG site -681 probe in the presence of 5- or 10-fold excess of methylated or unmethylated CpG site -681 double-stranded primers as well as Sp1 consensus oligonucleotides, and reactions were resolved on a 5% acrylamide gel. Band at the top of the gel represents Sp4/DNA complex while the free probe is at the bottom. Lane 1: CpG -681 probe alone; lane 2: CpG -681 probe + rSp4; lanes 3 & 4: CpG -681 probe+ rSp4 + 5x (3) or 10x (4) cold unmethylated CpG -681 competitor; lanes 5 & 6: CpG -681 probe+ rSp4 + 5x (5) or 10x (6) cold methylated CpG -681 competitor; lane 7: CpG -681 probe+ rSp4 + 10x cold Sp1 consensus. (B) Sp4 repressor activity at the -681 CpG element (model above). SN48, Neuro2A or SKN-SH cells were co-transfected with 1 ug of the pGL3P/-681CpG construct, Sp1, Sp4 or empty pTRIEX4 expression vector and pCMV β-gal. After 36h to 48h cells were lyzed and both luciferase and β-galactosidase activities were measured. Luciferase activity was normalized to β-galactosidase activity to correct for transfection efficiency. Data are presented as mean ±SEM of three independent experiments. ***p<0.001, **p<0.01, unpaired t-test. (C) Sp4 repressor activity is impaired by DNA methylation of 5-HT1A -684 CpG site. 25 bp oligomers containing methylated or unmethylated versions of the conserved promoter site were ligated in the KpnI/XhoI sites of pGL3P and transfected in Neuro2A cells along with the Sp4 or pTRIEX4 vector and pCMV β-gal. After 36h to 48h cells were lyzed and both luciferase and β-gal activities were measured. Luciferase activity was normalized to β-galactosidase activity to correct for transient transfection efficiency. Data presented as the ratio of pTRIEX4/pTRIEX4-Sp4 activities and is the mean ±SEM of four independent experiments. ***p<0.001 unpaired t-test.