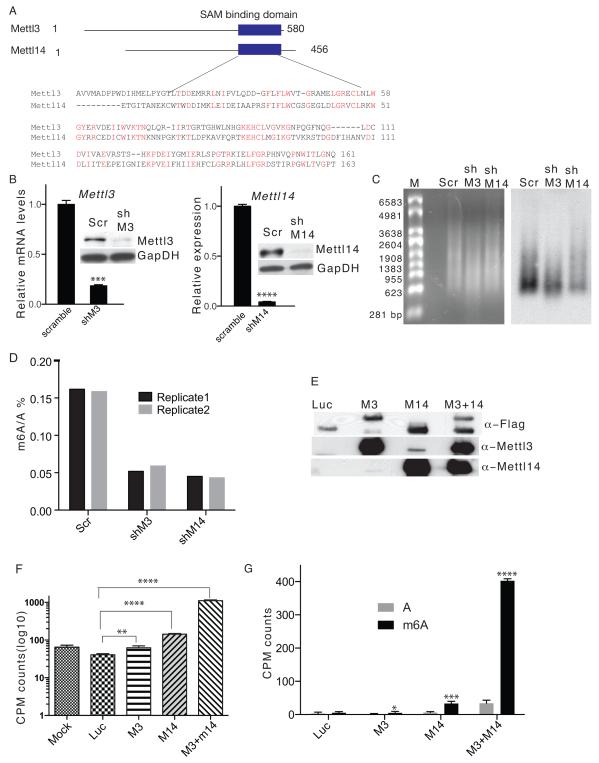
Figure 1. Mettl3 and Mettl14 are required for m6A formation in vitro and in vivo.
A. Schematic drawing showing predicted MTase domains of mouse Mettl3 (Accession: NP_062695.2) and Mettl14 (Accession: NP_964000.2) proteins. Numbers represent amino acid numbers. B. RT-qPCR (left) and western blot (right) showing Mettl3 (left panel) and Mettl14 (right panel) expression in mESC clones stably expressing shRNAs targeting each. GapDH serves as a loading control in the western blot. Scr: scramble; shM3, shRNA against Mettl3; shM14, shRNA against Mettl14. The original gel is shown in Supplementary Fig.5. C. Ethidium bromide staining (left panel) and m6A immunoblot (right panel) of DNA-free, rRNA-free Poly(A)+ RNA from Mettl3 kd, Mettl14 kd, and control cells. D. Measurement of percentage of m6A/A ratio by MS. E. Flag, Mettl3 or Mettl14 western immunoblots of proteins purified from lysates of HEK293 cells overexpressing flag-tagged Mettl3, Mettl14, or luciferase using M2 beads. Luc, luciferase; M3, Mettl3; M14, Mettl14. The original gel is shown in Supplementary Fig.5. F. CPM counts of RNA probes extracted after an in vitro methylation assay. G. CPM counts of excised m6A spots from digested RNA probes used in the methylation assay. Error bars from panels B and F-G represent means ± SEM from 3 separate experiments, expect M3 in panel G where n=7. One-tailed Student’s t-test, *P< 0.05, **P < 0.01, ***P < 0.001, ****P<0.0001 vs. scramble control. Scr: scramble control.