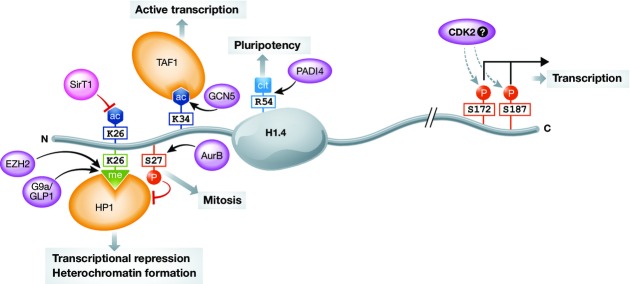
Figure 3. H1.4 modifications and their cellular functions.
Among the various modifications of linker histones, only few were characterized with site-specific antibodies. Mostly modifications on the subtype H1.4 have been characterized. H1.4 is methylated at K26, which is catalysed by G9a/GLP1 and potentially also by EZH2 120,134. This methylation provides a binding platform for HP1 and is thus linked to transcriptional repression and heterochromatin formation. Phosphorylation of S27 inhibits binding of HP1 to K26me 72. The C-terminal phosphorylations S172p and S187p are present on H1.4 also in interphase and have been linked to active transcription 116. H1.4K34ac is set by the acetyltransferase GCN5 and is enriched at active transcription start sites. It can positively regulate transcription by (i) recruiting the bromodomain-containing TAF1 subunit of the TFIID transcription complex and (ii) increasing H1 mobility 122. The conversion of R54 to citrulline by PADI4 has been shown to occur on several mouse H1 subtypes, among them H1.4. This modification in a DNA-binding site results in eviction of H1 and global chromatin decondensation in pluripotent cells. p: phosphorylation, ac: acetylation, me: methylation, cit: citrullination, violet: enzymes, orange: readers.