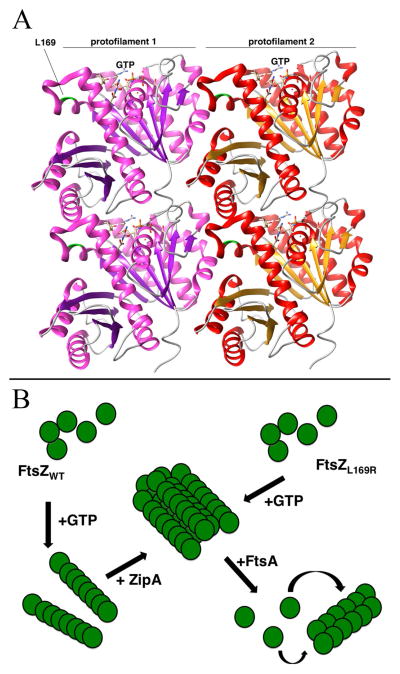
Figure 7.
Models for the effects of the L169R lesion on FtsZ protofilament interactions. (A) Potential structure of an FtsZ double protofilament, highlighting the position of L169 near the lateral interaction surface between two protofilaments and distal from the GTP binding site that is near the longitudinal interaction surface. The crystal structures from Pseudomonas FtsZ were manipulated with the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera) developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). The protofilament alignment was based on the atomic structures of FtsZ protofilaments (Li et al., 2013). (B) Scheme to compare assembly of FtsZWT or FtsZL169R into higher-order structures at the FtsZ ring. Upon binding to GTP, FtsZWT assembles into single protofilaments that are then bundled by the action of ZipA and Zap proteins (not shown). In contrast, upon binding to GTP, FtsZL169R assembles into protofilament bundles, reducing the need for additional bundling proteins. In both cases, FtsA acts as a counterbalance to FtsZ protofilament bundling, perhaps by destabilizing protofilament bundles, This putative de-bundling activity of FtsA normally inactivates FtsZ rings, but more highly bundled FtsZL169R rings are resistant.