Abstract
Although cell-free directed evolution methods have been used to engineer proteins for nearly two decades, selections on more complex phenotypes have largely remained in the domain of cell-based engineering approaches. Here, we review recent conceptual advances that now enable in vitro display of multimeric proteins, integral membrane proteins, and proteins with an expanded amino acid repertoire. Additionally, we discuss methodological improvements that have enhanced the accessibility, efficiency, and robustness of cell-free approaches. Coupling these advances with the in vitro advantages of creating exceptionally large libraries and precisely controlling all experimental conditions, cell-free directed evolution is poised to contribute significantly to our understanding and engineering of more complex protein phenotypes.
Introduction
Directed evolution is a powerful tool for tailoring biomolecular properties. For applications involving proteins, the approach requires a one-to-one mapping of phenotype and genotype, since selections or screens are performed on the proteins themselves, but amplification and identification of the desired sequences need to be performed at the genetic level. This phenotype-genotype linkage is easily achieved in cells, which can naturally compartmentalize individual DNA sequences and, through transcription and translation, the corresponding proteins. However, the use of cells significantly reduces the size of libraries that can be sampled, which can be vital, particularly when biomolecular functions are evolved completely de novo; additionally, cell-based directed evolution limits the scope of buffers, solvents, and temperatures that can be used since cell viability must be maintained.
In 1997, the first truly cell-free directed protein evolution platforms – mRNA display [1] and ribosome display [2] – were developed as iterative procedures that replicate the natural evolutionary processes of selection and amplification (Figure 1). The field of cell-free protein engineering has rapidly expanded over the past two decades, with notable advances in the use of defined translation mixtures [3], engineering of cell-toxic or aggregation-prone proteins [4], broadening selection and screening assays [5], and optimization of selection conditions and parameters [6]. By leveraging these advances and the unique evolutionary context provided by truly cell-free selections, researchers have also begun to investigate the origins of life [7]; in the context of structural biology, cell-free directed evolution provides a means for studying how primordial biomolecules evolved to adopt three-dimensional conformations that engendered novel functions [8*]. Many well-established applications of cell-free protein engineering approaches are comprehensively reviewed elsewhere [9,10]. Here, we focus on conceptual and methodological advances in cell-free directed protein evolution over the past few years, and we offer our perspective on the promising future directions of the field.
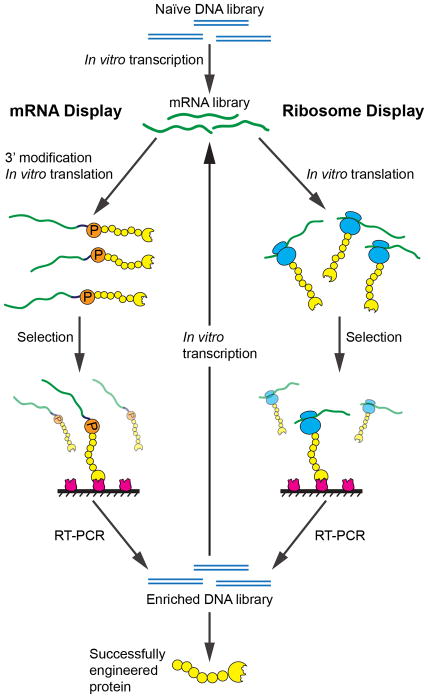
Figure 1. In vitro selection schemes for mRNA display and ribosome display.
A naïve DNA library is first transcribed into mRNA, for use in either mRNA display or ribosome display. In mRNA display, 3′ puromycin ligation and in vitro translation yield display particles comprised of mRNA and the corresponding translated protein, physically linked by puromycin. In ribosome display, in vitro translation of the mRNA directly yields the display particles, consisting of the mRNA, a stalled ribosome, and the corresponding translated protein. In each case, the display particles are then subjected to a selection assay, such as binding to an immobilized target. The selected particles undergo RT-PCR to yield an enriched DNA library. This enriched library is either transcribed again to go through subsequent in vitro display rounds or analyzed to obtain successfully engineered proteins.
Conceptual advances that have expanded the scope of selectable molecular phenotypes
Some of the most common phenotypes that cell-free protein engineers seek to create include high-affinity binding against cellular targets (ideally with high target specificity), more efficient biocatalysis (often paired with improved or altered substrate specificity), and higher protein stability under harsh solvent conditions. Such phenotypes have typically been evolved from monomeric, water-soluble protein templates. While the experimental focus on monomeric proteins can be attributed to the fact that most standard cell-free display methods fail to produce physical gene-protein linkages beyond a 1:1 stoichiometric ratio, the focus on water-soluble proteins relates to the poor in vitro expression and display properties of membrane proteins. Additionally, the cell-free evolutionary process has generally relied on changes in the amino acid sequence that could naturally occur in cells, but unnatural amino acids in the context of cell-free directed evolution could improve the functionality of engineered proteins. Here, we review recent conceptual advances in cell-free directed evolution that now enable monovalent display of multimeric proteins, multivalent display of monomeric proteins, display of integral membrane proteins, and incorporation of unnatural amino acids into displayed polypeptides (Figure 2).
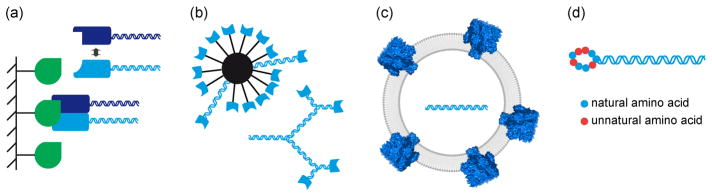
Figure 2. Schematic overview of recent conceptual advances in cell-free macromolecular display.
Novel or improved display particles and experimental designs expand the scope of protein phenotypes selectable by in vitro display methods. (a) Monovalent display of multimeric proteins [13**,16]. (b) Multivalent display of monomeric proteins [22,23*,25]. (c) Liposome display of integral membrane proteins [28*]. (d) mRNA display with incorporation of unnatural amino acids [33,34,38**].
Monovalent display of multimeric proteins
Native protein function often relies on multimeric structural organization. Antibodies represent one of the most common scaffolds in the protein engineering field, and displaying their hetero-oligomeric binding-competent fold is challenging [11]. Because all standard fully in vitro display systems fail to simultaneously encode and display multi-chain proteins, de novo combinatorial antibody discovery mostly relies on display methods that offer efficient cell-based chain-linking at the expense of throughput [12].
Recently, two groups have published completely cell-free affinity selection methods for Fab antibody fragments (Figure 2a). Both methods exploit the spontaneous assembly of the heavy chain (HC) and light chain (LC) into a functional, heterodimeric Fab fragment. In the method described by Sumida et al., two mRNA display sub-libraries, each displaying either HC or LC, are brought together to form a complete Fab fragment display library [13**]. After affinity selection, they perform gene-linking in vitro compartmentalization (IVC) PCR to physically link and then amplify the HC and LC gene pairs encoding the binding-competent heterodimeric Fab fragments. Importantly, and in contrast to similar earlier reports [14,15], the step that defines the upper limit of screenable library size (DNA linking by IVC; < 1010 individual compartments per mL of emulsion) is performed after affinity selection, so the full sampling power of mRNA display remains accessible. Tackling the same problem from a different angle, Stafford et al. used a modified ribosome display method for the selection of multi-chain Fab fragments [16]. In this approach, only one of the two Fab chains (either HC or LC) is mRNA-encoded and displayed on the ribosome during a single selection round, while the other protein chain is not presented in a display format but instead simply binds to the first protein chain to form a heterodimeric Fab fragment. The complete set of genes encoding binding-competent Fab fragments is then enriched by alternating the mRNA-encoded chain between HC and LC over multiple rounds of affinity selection.
Multivalent display of monomeric proteins
Display methods that tether multiple (on the order of millions) copies of a protein of interest (POI) to a single selection particle are formidable tools for affinity engineering of protein binders and for selecting very weak signals that only become detectable when multiple copies of the POI can contribute to the phenotype. For most applications, multivalent display particles are prepared using in vivo systems that tether multiple copies of a POI to the surface of either phage or whole cells. Making use of cellular biosynthesis, multivalency is easily achieved by genetically fusing the POI to a host protein that is abundantly expressed on the cell or phage surface [17–20].
The great versatility of cell-based multivalent selection technology has prompted strong interest in developing completely in vitro analogs that offer greater experimental flexibility (Figure 2b). The most common approach to mimicking cell-based multivalency in vitro is linking the genes and proteins to microbead surfaces by IVC [21]. Notable recent developments in this area include the use of microfluidic technologies to generate monodisperse droplets, preparation of microbeads displaying much larger numbers of proteins and DNA, and creating stronger linkages of the biomolecules to the beads [22,23*]. With these improvements, in vitro multivalent display particles now rival their cell-based counterparts in terms of valency but also allow control of their biomolecular composition to a degree not possible with cellular surfaces.
Additionally, exciting new bead-free approaches for multivalent display using engineered DNA scaffolds have been recently reported. To take advantage of avidity effects during affinity panning, Kaltenbach et al. have developed SNAP dendrimers consisting of dendrimer-like DNA templates covalently bound to POIs by the SNAP tag [24] at predefined valencies [25]. In model selections using SNAP dendrimers optimized for both DNA template geometry and display valency, they demonstrate 5- and 25-fold increases in particle recovery and enrichment, respectively, compared to monovalent display constructs. During early rounds of affinity selection, these effects could facilitate the recovery of more diverse sets of binders, including those that have low affinity but are still functionally interesting.
Display of integral membrane proteins
In contrast to water-soluble proteins, the distinct physicochemical properties of integral membrane proteins (IMP) have made them recalcitrant targets for combinatorial engineering. Standard in vitro display systems fail to properly accommodate the hydrophobic nature of IMPs; therefore, successful directed evolution of IMPs has relied on cell-based display systems [26,27]. Recently, Fujii et al. have reported a liposome display method (Figure 2c) that enables cell-free evolution of IMPs [28*]. The method combines in vitro transcription/translation (IVTT) with compartmentalization in giant unilamellar vesicles, resulting in the display of the IMP in the phospholipid bilayer of the liposome. Using the homo-oligomeric α-hemolysin as a template, the authors evolved a variant showing 30-fold higher pore-forming activity than wild type. The field of in vitro IMP evolution should benefit greatly from the rapid methodological advancements in the field of preparative IMP production using IVTT [29], which should pave the way for cell-free directed evolution of more complex IMPs.
Incorporation of unnatural amino acids
Incorporating unnatural amino acids into polypeptides can create completely novel phenotypes not found in nature. While the seminal work in this large research field is rooted in cell-based biosynthesis, unnatural amino acids have now become extremely useful building blocks for biomolecular engineering using in vitro display systems [30–32]. In fact, due to both the independence from cellular metabolic constraints and the high customizability of IVTT, cell-free display systems may open up enormous opportunities for selecting completely novel bioactive macromolecules composed of mostly unnatural amino acids.
The ribosomal production, display, and selection of peptide-like macrocycles represents one of the most exciting and promising research areas for in vitro display (Figure 2d). The key challenge to success is to achieve efficient biosynthesis using non-peptidogenic amino acids. To date, two different display methods have been published. While both methods share the puromycin-based mechanism of mRNA display to achieve covalent genotype-phenotype coupling, they use different strategies for reprogramming the genetic code and customizing the translation machinery. Yamagishi et al. reported selection of a macrocycle inhibitor of the E6AP ubiquitin ligase using a display system referred to as random nonstandard peptides integrated discovery (RaPID) [33]. Schlippe et al. successfully carried out selections of highly unnatural macrocyclic peptides against thrombin [34]. Both groups recovered binders with low nanomolar affinity mediated by the unnatural amino acids, demonstrating successful selection of highly non-peptidogenic macrocycles from gene-encoded libraries by in vitro display. Furthermore, the RaPID system has yielded potent macrocyclic inhibitors of a microbial multidrug transporter. These macrocycles successfully served as co-crystallization ligands to elucidate key molecular determinants of drug efflux pumping and molecular modes of transport inhibition [35**]. Macrocycles discovered by in vitro display methods may represent very interesting drug candidates, because they occupy a region of chemical and functional space that is positioned between the two extremes of small molecule drugs and large biologics.
Using ribosome display of peptides and incorporating only a single type of unnatural amino acid – the photo-responsive azobenzoyl-lysine – peptides could be evolved that bind to streptavidin in a light-dependent fashion by cis-trans isomerization of the azobenzoyl group [36]. A different type of photoactive peptide, one that emits increased fluorescence upon binding to calmodulin, was selected from a library in which the peptides contained a fluorogenic amino acid [37]. Lastly, cell-free affinity selection of multivalent glycopeptides was achieved using the unnatural amino acid homopropargylglycine, which enables attachment of glycans via “click” chemistry, and it was shown that nanomolar binding affinity to an antibody paratope was achieved through the sugar moiety [38**]. These examples demonstrate that unnatural amino acids can be used as tools for post-translationally modifying phenotypes of displayed peptides, and these more complex phenotypes will enable more sophisticated selections.
Methodological advances that have improved accessibility, efficiency, and robustness of cell-free approaches
Beyond conceptual innovations widening the breadth of problems that in vitro selections can address, recent methodological advances have enhanced the accessibility, efficiency, and robustness of existing cell-free selection schemes (Figure 3). These improvements set the stage for reliable methods that researchers can readily implement to carry out cell-free directed evolution.
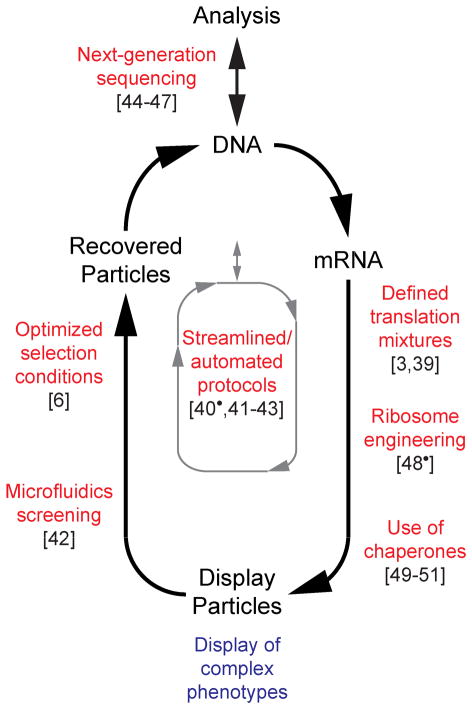
Figure 3. General flow diagram of cell-free directed evolution with notable recent advances highlighted.
During each round of evolution (black), the DNA library is transcribed to mRNA and then translated under specific conditions to create selectable particles that link genotype and phenotype. These particles undergo a selection or screening step, and recovered particles are converted back into an enriched DNA library, which can be further analyzed or directly input into a new round. Conceptual advances (blue) focus on improving the display particles to allow selection of complex molecular phenotypes, as more specifically illustrated in Figure 2. Methodological advances (red) improve the accessibility, efficiency, and robustness of the distinct processes involved in in vitro directed evolution, and can generally reduce the time required for successful evolution (grey).
Accessibility of cell-free technologies to new researchers remains a challenge, since complete cell-free directed evolution kits are not currently commercially available (e.g., in contrast to phage display kits) and existing approaches are generally technically demanding. However, new products that encourage entry into the field while also enhancing the quality of libraries for established protein engineers are emerging. For example, the PURE system [3,39] is a fully defined IVTT system with a high concentration of ribosomes and low levels of detrimental nucleases and proteases, thus yielding more complex libraries of display particles. Additionally, streamlined protocols are being developed to simplify cell-free evolution [40*,41].
More efficient screening and selection procedures can help accelerate cell-free laboratory evolution. Mathematical modeling has been used to uncover insights that can optimize cell-free selection experiments to more effectively recover high-affinity-binding proteins [6]. Considering the inherently high sampling power of in vitro display systems, experimental throughput is often limited by single-clone screening of enriched library pools. The latest advances in microfluidic droplet generation with on-chip sorting allow for integrated parsing of 2,000 clones per second [42]. Cell-free selections also lend themselves to automation because of the repetitive nature of cycling through rounds; establishing clearly defined steps with tunable parameters removes a labor-intensive barrier and makes the selection more efficient [43]. Additionally, next-generation sequencing (NGS) enables data-driven analytics to better inform selection schemes [44], reducing the number of rounds necessary for successful selections [45,46]. In stark contrast to conventional selection schemes involving multiple selection rounds and sequencing of only highly-enriched clones, Olson et al. demonstrated successful cell-free evolution by performing just one round of mRNA display followed by NGS analysis [47].
The robustness of system components is critical to the success of cell-free evolution platforms because this helps to minimize losses in library diversity during directed evolution rounds and post-evolution screening. A method known as iSAT could enable engineering of the ribosome for greater functionality [48*], improving translation in all cell-free evolution schemes. Introduction of molecular chaperones to cell-free translation systems increases protein yield [49] and prevents aggregation of proteins and their mutants by promoting proper folding [50]; similarly, nanogel artificial chaperones are emerging as an alternative means to help mitigate library losses [51].
Conclusion and Outlook
Combinatorial cell-free protein engineering continues to be an active, quickly-developing field, providing the research community established and novel tools for creating and improving biomolecular function. When biomolecular engineering projects rely on either sampling the largest possible libraries or exerting full control over the experimental selection conditions, in vitro display methods are clearly favored over cell-based schemes. Here we have reviewed recent work highlighting the expanding scope of cell-free biomolecular display. Conceptually, both the increasingly efficient biosynthetic incorporation of unnatural amino acids into POIs and the possibility to display multimeric POIs without compromising sampling power hold great promise for expanding the scope of selectable protein phenotypes. Technically, notable advances include the use of NGS to analyze much larger datasets of selected clones, the increase in control over the selection milieu due to fully defined IVTT systems, and the multitude of protocol improvements and simplifications of in vitro display methods.
Looking forward, several advances are likely to have a profound impact on biomolecular combinatorial engineering. Reprogramming the biosynthetic machinery with expanded sets of both unnatural nucleotides [52] and amino acids [33,34] will lead to an exponential increase in the (bio)chemical space that can be sampled. Using NGS technology to more comprehensively decode and analyze sets of selected genes will improve experimental design and success. The development of microfluidic droplet generation platforms as powerful alternatives to multiwell microplates will drastically increase the throughput for one-by-one biochemical screening of evolved proteins [42,53–55]. Furthermore, in vitro directed evolution will play an integral role in optimizing networks of biomolecular interactions in cell-free synthetic biology [56,57]. The emergence of these new conceptual and methodological advances in cell-free directed evolution will enable biomolecular engineers to more rapidly gain fundamental insights into structure-function relationships – especially in structural studies that benefit from non-native protein folds [8*] or biochemical properties [35**] – and to create better biomolecules for biotechnological and biomedical applications.
Highlights.
Advances in cell-free evolution allow for more sophisticated protein engineering.
New display formats enable evolution of multimeric and integral membrane proteins.
Incorporation of unnatural amino acids expands chemical repertoire in vitro.
Technical enhancements improve method accessibility, efficiency, and robustness.
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA179180) and the National Science Foundation (CBET-1055231) to C.A.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci U S A. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci U S A. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu Y, Kanamori T, Ueda T. Protein synthesis by pure translation systems. Methods. 2005;36:299–304. doi: 10.1016/j.ymeth.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Schimmele B, Grafe N, Plückthun A. Ribosome display of mammalian receptor domains. Protein Eng Des Sel. 2005;18:285–294. doi: 10.1093/protein/gzi030. [DOI] [PubMed] [Google Scholar]
- 5.Seelig B, Szostak JW. Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature. 2007;448:828–831. doi: 10.1038/nature06032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zahnd C, Sarkar CA, Plückthun A. Computational analysis of off-rate selection experiments to optimize affinity maturation by directed evolution. Protein Eng Des Sel. 2010;23:175–184. doi: 10.1093/protein/gzp087. [DOI] [PubMed] [Google Scholar]
- 7.Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Chao FA, Morelli A, Haugner JC, 3rd, Churchfield L, Hagmann LN, Shi L, Masterson LR, Sarangi R, Veglia G, Seelig B. Structure and dynamics of a primordial catalytic fold generated by in vitro evolution. Nat Chem Biol. 2013;9:81–83. doi: 10.1038/nchembio.1138. An RNA ligase was evolved from an entirely non-enzymatic library of proteins containing DNA-binding zinc finger motifs. Structural analysis of this enzyme revealed a novel fold that resulted from evolution in the absence of selection pressure to maintain the original function of DNA binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldsmith M, Tawfik DS. Directed enzyme evolution: beyond the low-hanging fruit. Curr Opin Struct Biol. 2012;22:406–412. doi: 10.1016/j.sbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Lane MD, Seelig B. Advances in the directed evolution of proteins. Curr Opin Chem Biol. 2014;22:129–136. doi: 10.1016/j.cbpa.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams JJ, Sidhu SS. Synthetic antibody technologies. Curr Opin Struct Biol. 2014;24:1–9. doi: 10.1016/j.sbi.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19:4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Sumida T, Yanagawa H, Doi N. In vitro selection of fab fragments by mRNA display and gene-linking emulsion PCR. Journal of Nucleic Acids. 2012;2012:371–379. doi: 10.1155/2012/371379. This method optimally combines mRNA display and IVC to allow affinity selection of heterodimeric Fab fragments. The key to fully preserving the library sampling power of mRNA display is to perform the library-limiting step (the IVC-promoted linking of the two genes encoding the heterodimer) after affinity panning. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumida T, Doi N, Yanagawa H. Bicistronic DNA display for in vitro selection of Fab fragments. Nucleic Acids Res. 2009;37:e147. doi: 10.1093/nar/gkp776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sepp A, Griffiths A. Cell-free selection of domain antibodies by in vitro compartmentalization. Methods Mol Biol. 2012;911:183–198. doi: 10.1007/978-1-61779-968-6_12. [DOI] [PubMed] [Google Scholar]
- 16.Stafford RL, Matsumoto ML, Yin G, Cai Q, Fung JJ, Stephenson H, Gill A, You M, Lin S-H, Wang WD, et al. In vitro Fab display: a cell-free system for IgG discovery. Protein Eng Des Sel. 2014;27:97–109. doi: 10.1093/protein/gzu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 18.Freudl R, MacIntyre S, Degen M, Henning U. Cell surface exposure of the outer membrane protein OmpA of Escherichia coli K-12. J Mol Biol. 1986;188:491–494. doi: 10.1016/0022-2836(86)90171-3. [DOI] [PubMed] [Google Scholar]
- 19.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 20.Ho M, Nagata S, Pastan I. Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc Natl Acad Sci U S A. 2006;103:9637–9642. doi: 10.1073/pnas.0603653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sepp A, Tawfik DS, Griffiths AD. Microbead display by in vitro compartmentalisation: selection for binding using flow cytometry. FEBS Lett. 2002;532:455–458. doi: 10.1016/s0014-5793(02)03740-7. [DOI] [PubMed] [Google Scholar]
- 22.Paul S, Stang A, Lennartz K, Tenbusch M, Überla K. Selection of a T7 promoter mutant with enhanced in vitro activity by a novel multi-copy bead display approach for in vitro evolution. Nucleic Acids Res. 2013;41:e29. doi: 10.1093/nar/gks940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Diamante L, Gatti-Lafranconi P, Schaerli Y, Hollfelder F. In vitro affinity screening of protein and peptide binders by megavalent bead surface display. Protein Eng Des Sel. 2013;26:713–724. doi: 10.1093/protein/gzt039. This megavalent display method (up to 106 displayed molecules per bead) provides robust control over the number of displayed peptides and DNA molecules per bead. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 25.Kaltenbach M, Stein V, Hollfelder F. SNAP dendrimers: multivalent protein display on dendrimer-like DNA for directed evolution. Chem Bio Chem. 2011;12:2208–2216. doi: 10.1002/cbic.201100240. [DOI] [PubMed] [Google Scholar]
- 26.Sarkar CA, Dodevski I, Kenig M, Dudli S, Mohr A, Hermans E, Plückthun A. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci U S A. 2008;105:14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dodevski I, Plückthun A. Evolution of three human GPCRs for higher expression and stability. J Mol Biol. 2011;408:599–615. doi: 10.1016/j.jmb.2011.02.051. [DOI] [PubMed] [Google Scholar]
- 28*.Fujii S, Matsuura T, Sunami T, Kazuta Y, Yomo T. In vitro evolution of α-hemolysin using a liposome display. Proc Natl Acad Sci U S A. 2013;110:16796–16801. doi: 10.1073/pnas.1314585110. Integral membrane proteins are recalcitrant targets for in vitro display. By performing cell-free protein synthesis of an α-hemolysin library in giant unilamellar vesicles, the authors successfully display and select mutants showing strongly increased channel-forming properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roos C, Kai L, Haberstock S, Proverbio D, Ghoshdastider U, Ma Y, Filipek S, Wang X, Dotsch V, Bernhard F. High-level cell-free production of membrane proteins with nanodiscs. Methods Mol Biol. 2014;1118:109–130. doi: 10.1007/978-1-62703-782-2_7. [DOI] [PubMed] [Google Scholar]
- 30.Bhat A, Roberts LR, Dwyer JJ. Lead discovery and optimization strategies for peptide macrocycles. European Journal of Medicinal Chemistry. 2014 doi: 10.1016/j.ejmech.2014.07.083. [DOI] [PubMed] [Google Scholar]
- 31.Josephson K, Ricardo A, Szostak JW. mRNA display: from basic principles to macrocycle drug discovery. Drug Discov Today. 2014;19:388–399. doi: 10.1016/j.drudis.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 32.Passioura T, Katoh T, Goto Y, Suga H. Selection-based discovery of druglike macrocyclic peptides. Annu Rev Biochem. 2014;83:727–752. doi: 10.1146/annurev-biochem-060713-035456. [DOI] [PubMed] [Google Scholar]
- 33.Yamagishi Y, Shoji I, Miyagawa S, Kawakami T, Katoh T, Goto Y, Suga H. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem Biol. 2011;18:1562–1570. doi: 10.1016/j.chembiol.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Schlippe YVG, Hartman MCT, Josephson K, Szostak JW. In vitro selection of highly modified cyclic peptides that act as tight binding inhibitors. J Am Chem Soc. 2012;134:10469–10477. doi: 10.1021/ja301017y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Tanaka Y, Hipolito CJ, Maturana AD, Ito K, Kuroda T, Higuchi T, Katoh T, Kato HE, Hattori M, Kumazaki K, et al. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature. 2013;496:247–251. doi: 10.1038/nature12014. Macrocyclic peptides are selected in vitro to inhibit a bacterial multidrug transporter and serve as co-crystallization ligands for solving the crystal structure of the inhibited protein conformation. [DOI] [PubMed] [Google Scholar]
- 36.Liu M, Tada S, Ito M, Abe H, Ito Y. In vitro selection of a photo-responsive peptide aptamer using ribosome display. Chem Commun (Camb) 2012;48:11871–11873. doi: 10.1039/c2cc36618e. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Uzawa T, Tochio N, Hamatsu J, Hirano Y, Tada S, Saneyoshi H, Kigawa T, Hayashi N, Ito Y, et al. A fluorogenic peptide probe developed by in vitro selection using tRNA carrying a fluorogenic amino acid. Chem Commun (Camb) 2014;50:2962–2964. doi: 10.1039/c3cc47624c. [DOI] [PubMed] [Google Scholar]
- 38**.Horiya S, Bailey JK, Temme JS, Guillen Schlippe YV, Krauss IJ. Directed evolution of multivalent glycopeptides tightly recognized by HIV antibody 2G12. J Am Chem Soc. 2014;136:5407–5415. doi: 10.1021/ja500678v. This in vitro method addresses the challenge of engineering artificial scaffolds that precisely mimic natural glycan clustering and may serve as a powerful tool for discovering peptide backbones for highly antigenic presentation of glycans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittaker JW. Cell-free protein synthesis: the state of the art. Biotechnol Lett. 2013;35:143–152. doi: 10.1007/s10529-012-1075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Ishizawa T, Kawakami T, Reid PC, Murakami H. TRAP display: a high-speed selection method for the generation of functional polypeptides. J Am Chem Soc. 2013;135:5433–5440. doi: 10.1021/ja312579u. By combining transcription, translation, and puromycin incorporation into a single procedural step, the time required per round of mRNA display is drastically reduced from 2–3 days to 2–3 hours. [DOI] [PubMed] [Google Scholar]
- 41.Barendt PA, Ng DT, McQuade CN, Sarkar CA. Streamlined protocol for mRNA display. ACS Comb Sci. 2013;15:77–81. doi: 10.1021/co300135r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fallah-Araghi A, Baret J-C, Ryckelynck M, Griffiths AD. A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab on a Chip. 2012;12:882–891. doi: 10.1039/c2lc21035e. [DOI] [PubMed] [Google Scholar]
- 43.Doshi R, Chen BR, Vibat CRT, Huang N, Lee C-W, Chang G. In vitro nanobody discovery for integral membrane protein targets. Sci Rep. 2014;4:6760. doi: 10.1038/srep06760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larman HB, Xu GJ, Pavlova NN, Elledge SJ. Construction of a rationally designed antibody platform for sequencing-assisted selection. Proc Natl Acad Sci U S A. 2012;109:18523–18528. doi: 10.1073/pnas.1215549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravn U, Gueneau F, Baerlocher L, Osteras M, Desmurs M, Malinge P, Magistrelli G, Farinelli L, Kosco-Vilbois MH, Fischer N. By-passing in vitro screening–next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res. 2010;38:e193. doi: 10.1093/nar/gkq789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heyduk E, Heyduk T. Ribosome display enhanced by next generation sequencing: a tool to identify antibody-specific peptide ligands. Anal Biochem. 2014;464:73–82. doi: 10.1016/j.ab.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 47.Olson CA, Nie J, Diep J, Al-Shyoukh I, Takahashi TT, Al-Mawsawi LQ, Bolin JM, Elwell AL, Swanson S, Stewart R, et al. Single-round, multiplexed antibody mimetic design through mRNA display. Angew Chem Int Ed Engl. 2012;51:12449–12453. doi: 10.1002/anie.201207005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Jewett MC, Fritz BR, Timmerman LE, Church GM. In vitro integration of ribosomal RNA synthesis, ribosome assembly, and translation. Mol Syst Biol. 2013;9:678. doi: 10.1038/msb.2013.31. This in vitro method for Escherichia coli ribosomal reconstitution integrates rRNA synthesis, ribosome assembly, and protein translation into a single step. This procedure can enable cell-free evolution approaches to better understand and engineer ribosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Gu L, Aach J, Church GM. Improved cell-free RNA and protein synthesis system. PLoS One. 2014;9:e106232. doi: 10.1371/journal.pone.0106232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niwa T, Kanamori T, Ueda T, Taguchi H. Global analysis of chaperone effects using a reconstituted cell-free translation system. Proc Natl Acad Sci U S A. 2012;109:8937–8942. doi: 10.1073/pnas.1201380109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki Y, Asayama W, Niwa T, Sawada S, Ueda T, Taguchi H, Akiyoshi K. Amphiphilic polysaccharide nanogels as artificial chaperones in cell-free protein synthesis. Macromol Biosci. 2011;11:814–820. doi: 10.1002/mabi.201000457. [DOI] [PubMed] [Google Scholar]
- 52.Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa IR, Jr, Romesberg FE. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509:385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo MT, Rotem A, Heyman JA, Weitz DA. Droplet microfluidics for high-throughput biological assays. Lab Chip. 2012;12:2146–2155. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 54.Miller OJ, El Harrak A, Mangeat T, Baret JC, Frenz L, El Debs B, Mayot E, Samuels ML, Rooney EK, Dieu P, et al. High-resolution dose-response screening using droplet-based microfluidics. Proc Natl Acad Sci U S A. 2012;109:378–383. doi: 10.1073/pnas.1113324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El Debs B, Utharala R, Balyasnikova IV, Griffiths AD, Merten CA. Functional single-cell hybridoma screening using droplet-based microfluidics. Proc Natl Acad Sci U S A. 2012;109:11570–11575. doi: 10.1073/pnas.1204514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodgman CE, Jewett MC. Cell-free synthetic biology: thinking outside the cell. Metab Eng. 2012;14:261–269. doi: 10.1016/j.ymben.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith MT, Wilding KM, Hunt JM, Bennett AM, Bundy BC. The emerging age of cell-free synthetic biology. FEBS Lett. 2014;588:2755–2761. doi: 10.1016/j.febslet.2014.05.062. [DOI] [PubMed] [Google Scholar]