Abstract
In bacterial RNA polymerase (RNAP), the bridge helix and switch regions form an intricate network with the catalytic active centre and the main channel. These interactions are important for catalysis, hydrolysis and clamp domain movement. By targeting conserved residues in Escherichia coli RNAP, we are able to show that functions of these regions are differentially required during σ70-dependent and the contrasting σ54-dependent transcription activations and thus potentially underlie the key mechanistic differences between the two transcription paradigms. We further demonstrate that the transcription factor DksA directly regulates σ54-dependent activation both positively and negatively. This finding is consistent with the observed impacts of DksA on σ70-dependent promoters. DksA does not seem to significantly affect RNAP binding to a pre-melted promoter DNA but affects extensively activity at the stage of initial RNA synthesis on σ54-regulated promoters. Strikingly, removal of the σ54 Region I is sufficient to invert the action of DksA (from stimulation to inhibition or vice versa) at two test promoters. The RNAP mutants we generated also show a strong propensity to backtrack. These mutants increase the rate of transcript-hydrolysis cleavage to a level comparable to that seen in the Thermus aquaticus RNAP even in the absence of a non-complementary nucleotide. These novel phenotypes imply an important function of the bridge helix and switch regions as an anti-backtracking ratchet and an RNA hydrolysis regulator.
Abbreviations: RNAP, RNA polymerase; WT, wild type
Keywords: RNA polymerase, bridge helix, switch regions, DksA, RNA hydrolysis
Graphical abstract
Highlights
-
•
The bridge helix and switch regions form an intricate network in RNAP.
-
•
The σ70 and σ54 transcription systems differentially use this interaction network.
-
•
Transcription factor DksA and σ54 Region I also contribute to this network.
-
•
Disruption of this network enhances backtracking and intrinsic RNA hydrolysis.
Introduction
Multisubunit RNA polymerases (RNAPs) carry out the essential function of DNA transcription and are highly conserved in structure and function across bacteria, archaea and eukaryotes [1], [2], [3]. The bacterial RNAP holoenzyme consists of a catalytic core (α2ββ′ω or E) and a sigma factor (σ) required for promoter specific recognition [4], [5]. The two large β and β′ subunits form a crab claw-like structure, with the mobile β lobe, β′ clamp and β′ jaw forming the two pincers [6]. Part of the β′ clamp hinges onto the RNAP body via the switch regions SW and has been shown to swing open for DNA entry and remain closed during initiation and elongation, respectively [7]. Five segments of the switch regions control the β′ clamp movement, termed switches 1–5 (SW 1–5), and they are also a new class of antibiotics target sites [8], [9], [10]. Myxopyronin is one antibiotic that targets the switch regions [9], [11]. Structural analyses indicate that myxopyronin makes direct contacts with SW 1, 2, 4 and 5 [9]. Residues of SW 1–3 make direct contacts with the DNA template [8], [12], [13]. However, conformational changes of SW 1, 2, 4 and 5 are thought to promote the refolding of SW 3 [14]. These switches interact with one another and their conformational changes can be coupled to that of the bridge helix [8].
The bridge helix BH bifurcates the RNAP central cleft into a main DNA loading channel and a secondary channel [15]. As free nucleotide NTP is proposed to enter the secondary channel and binds to the “i + 1” site within the active centre for base pairing, the trigger loop TL locks in the correct NTP using its refolded helical conformation [16]. It has been proposed that the trigger helices kink the BH that subsequently shifts the nascent 3′-NMP from the “i + 1” site to the “i” site, driving the forward movement of the RNAP from a pre-translocated state to a post-translocated state [17]. The polymerisation can be reversed in cases of processive pyrophosphorolysis or intrinsic and assisted cleavages of the backtracked RNA [17], [18], [19], [20].
The activity and conformation of the RNAP active centre are subject to modulation from transcription factors bound to the secondary channel [17], [21], [22], [23], [24]. For instance, DksA presents two Asp residues at the tip of its coiled-coil domain to the active centre during stringent response [25]. It has been proposed that DksA amplifies the inhibitory effect of ppGpp and reduces the half-life of certain open promoter complexes, possibly by allosterically affecting SW and/or BH/TL [26]. The allosteric regulation of the distant SW regions is thought to be achieved via the DksA's coiled-coil tip and the BH/TL three-helical bundle, which in turn could affect clamp opening and DNA contacts [22]. Consistent with this proposal, DksA-suppressor mutations were found in the BH/TL and four of the five SW regions (SW 1–3 and 5) [26]. The reason we chose BH/SW residues outside of the established DksA-suppressor mutations is twofold: (i) we would like to examine the impact of DksA on σ54-regulated promoters, and (ii) we would like to assess the potential interactions between DksA and the σ54 Region I.
The catalytic dynamics of bacterial RNAP are defined by distinct multiple energetic states along the activation pathway. In Escherichia coli, the σ70 factor directs RNAP to the − 10 and − 35 elements and spontaneously isomerises the closed complex (RPC) to an open complex (RPO) via several intermediates (RPIs). Under stress conditions, many genes that are involved in pathogenicity, nitrogen assimilation and biofilm formation use the alternative σ54 factor [27], [28], [29], [30], [31]. σ54 binds to the − 12 and − 24 elements to form the RPC and imposes a high-energy barrier to prevent DNA melting by inhibitory action of its Region I (σ54RI [32], [33]). Hexameric AAA+ σ54 activators (such as the E. coli phage shock protein PspF and the nitrogen regulation protein NtrC) help transcend the energy barrier at the expense of ATP hydrolysis, leading to full DNA melting in the RPO [34], [35], [36]. These properties mimic some eukaryotic Pol II characteristics, and distinct RPIs of σ54-dependent transcription complexes can be efficiently captured by using non-hydrolysable ATP analogues bound to its activators [37], [38], [39], [40].
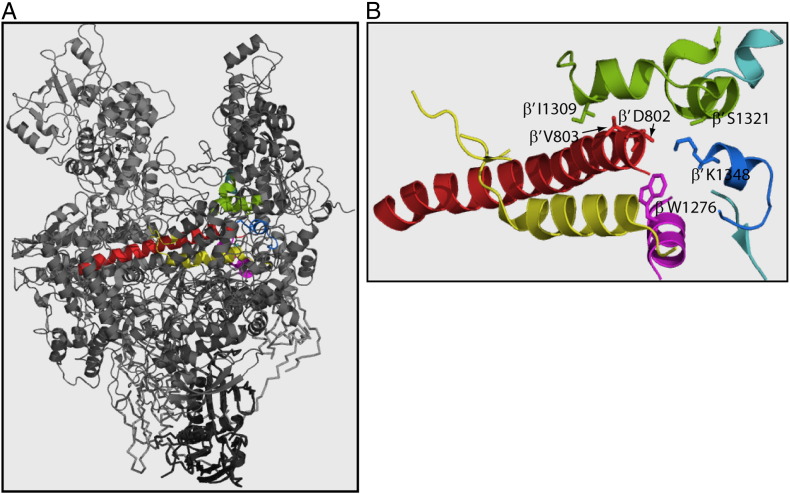
Seven residues in (or in close proximity to) the BH and SW regions were selected for this study based on their sequence conservation (Fig. S1). These residues are located near the active centre and the secondary channel (Fig. 1), and these could have potential impacts on RNA extension, hydrolysis and functions of transcription factors. The majority of these selected residues were mutated to an Ala to assess their side-chain interactions. Position S1321 was further mutated to a Lys to assess the impact of pair-swapping with K1348 on myxopyronin binding [11]. The wild-type (WT) and mutant RNAPs were normalised based on β and β′ concentrations (Fig. S2) and then reconstituted as holoenzymes with purified sigma factors (σ70 or σ54). The BH and SW mutations in general are detrimental to RPO formation, abortive synthesis and transcription elongation. We find that DksA imposes both positive and negative effects on σ54-dependent promoters. We speculate that, aside from the intrinsic properties of the promoter DNA, DksA works synergistically with σ54 Region I to regulate certain activator-dependent transcription complexes. The RNAP mutants we generated stay preferentially in a backtracked state and can efficiently perform an intrinsic penultimate phosphodiester bond cleavage without further requirement of mismatching NMP or inorganic pyrophosphate. Once locked in the backtracked state, many of the mutants do not elongate upon the addition of the next NTP.
Fig. 1.
Residues of the bridge helix and switch regions selected for this study. (A) The bridge helix (red), SW 1 (green), SW 2 (cyan), N-terminus of SW 5 (blue), C-terminus of SW 3 (magenta) and trigger loop (yellow) are highlighted in the E. coli crystal structure (PDB entry 4IGC). (B) The BH and SW regions were zoomed in to show residues of interest.
Results
BH and SW residues impact on both RPO formation and initial RNA synthesis
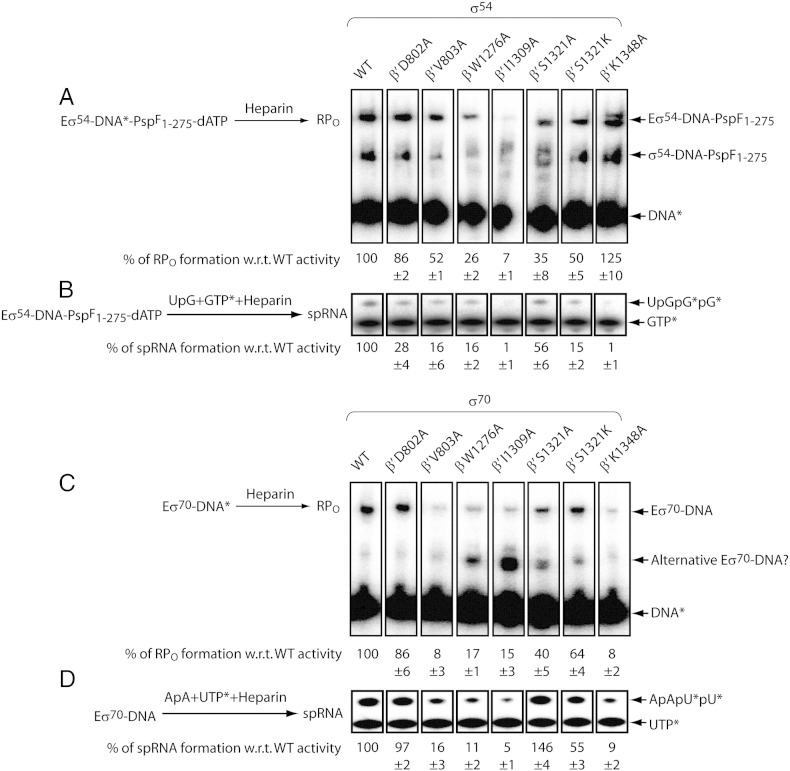
The BH and SW residues form an intricate network around the RNAP active centre, in particular, with the promoter DNA and the trigger loop. Disruption of such network could potentially lead to defects in RPO formation and abortive synthesis. To assess the σ54-dependent RPO formation, we assembled the Eσ54 holoenzymes with activator PspF1 -275 and activating nucleotide dATP on a test Sinorhizobium meliloti nifH linear promoter probe (harbouring a mismatch from − 10 to − 1 on the non-template strand to mimic the fully melted transcription bubble, − 10 − 1/WT) and subsequently challenged them with heparin. Several RNAP mutants tested showed modest (50–90% WT activity) to severe defects (below 50% WT activity) in RPO formation (Fig. 2A). In contrast, the β′ K1348A mutant is more efficient in generating RPO (125% WT activity; Fig. 2A). To test if these mutants could carry the RPO into initial RNA synthesis, we added a dinucleotide primer and 32P-GTP to the transcription complex. The dinucleotide UpG and 32P-GTP form the tetranucleotide UpGpGpG (as the small primed RNA) whose level correlates with that of a forced short RNA synthesis product (Fig. 2B). In the cases of the β′ D802A, β′ V803A, β′ 1321K and β′ K1348A mutants, the sufficient amount of heparin-resistant RPO did not ensure an equivalent level of abortive synthesis (compare Fig. 2B with 2A). The β′ I1309A mutant completely abolished abortive synthesis, primarily due to its loss of RPO formation (compare Fig. 2B with 2A). The abovementioned data suggest that some of the BH and SW residues contribute to the stability of σ54-dependent RPO and more importantly the rate of abortive synthesis.
Fig. 2.
The BH and SW mutants are defective in RPO formation and abortive synthesis. (A) The σ54-dependent RPO formation was assembled on a radiolabelled (*) nifH − 10−1/WT probe, challenged by heparin and analysed on a native gel. (B) The σ54-dependent abortive synthesis was assessed on the same DNA with a dinucleotide primer UpG and 32P-GTP to generate small primed RNA (spRNA; UpGpGpG). The σ70-dependent RPO formation (C) and abortive synthesis (D) were assessed on a radiolabelled lacUV5 − 10−1/WT probe. As σ70 does not usually form heparin-resistant complexes with DNA, the fast migrating band [in (c)] may represent an alternative form of Eσ70–DNA complexes. “w.r.t.” stands for “with respect to”.
In the σ70-dependent RPO formation studies, the Eσ70 holoenzymes were reconstituted on an E. coli lacUV5 − 10−1/WT linear promoter probe and challenged with heparin. In comparison to the σ54-dependent RPO formation studies, the β′ V803A and β′ K1348A mutants showed the most pronounced reduction in RPO formation, whereas the β′ I1309A and β′ S1321K mutants showed marginally improved activities (compare Fig. 2C with 2A). In comparison to the σ54-dependent abortive synthesis, the β′ D802A, β′ S1321A and β′ S1321K showed the most significant improvement (compare Fig. 2D with 2B). The β W1276A and β′ I1309A possessed very low binding and catalytic activities in both transcription systems. These data indicate that the BH and SW residues may determine different functions in the σ54 and σ70 transcription paradigms.
DksA significantly affects abortive synthesis but not RPO formation on four σ54-dependent promoters
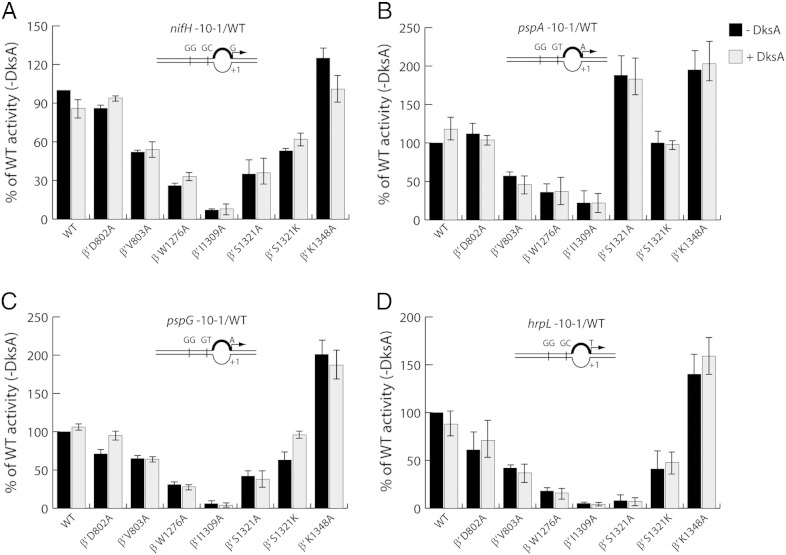
DksA can independently and directly regulate promoters transcribed by Eσ70 [41], [42]. However, in the case of σ54-dependent promoters, the effects of DksA on in vitro transcription are passive, as its addition does not significantly change the RPO lifetime or transcript level on a Pseudomonas putida Po promoter [43]. We suspect that the marked regulatory differences of DksA on the two transcription systems may involve intrinsic promoter properties. Thus, four linear σ54-dependent promoter probes were constructed: S. meliloti nifH (nitrogen fixation [32]), E. coli pspA and pspG (membrane stress response [44]) and Pseudomonas syringae hrpL (type III secretion system [45]), all harbouring a pre-melted transcription bubble from − 10 to − 1 (Fig. 3, diagrams). The RPO formation assays were performed as described above and analysed on native gels. In agreement with the passive model as previously proposed, the presence of DksA does not significantly change the stability of RPO on all four σ54-dependent promoters (Fig. 3). The ability of each RNAP mutant to form RPO on different promoters is generally conserved. The exception lies with the β′ S1321A mutant that showed significantly improved RPO formation on the pspA promoter over the other three promoters (compare Fig. 3).
Fig. 3.
DksA does not seem to significantly impact on σ54-dependent RPO formation. (A–D) Four σ54-dependent linear promoter probes (nifH, pspA, pspG and hrpL), all harbouring a mismatch from − 10 to − 1 on the non-template strand (thick lines in diagram), were used to generate heparin-resistant RPO with σ54, PspF1 -275 and dATP on native gels. The activity of each RNAP mutant in the absence and presence of DksA (black and grey bars, respectively) was expressed as a percentage of that of WT RNAP without DksA for each promoter. The − 24 and − 12 elements and the transcription start sites are indicated in the diagram. Each experiment was reproduced in triplicate. The error bars represent standard deviation.
As the rate of abortive synthesis was assessed, the first striking observation was that DksA had direct and differential impacts on all four σ54-dependent promoters (Fig. 4). The abortive synthesis with the WT RNAP was stimulated by DksA by more than 2-fold on the nifH promoter (Fig. 4A) and by nearly 50% on the pspG promoter (Fig. 4C) but inhibited by approximately 30% on the hrpL promoter (Fig. 4D). The BH and SW mutants reacted to σ54-dependent promoters to different extents, with the β′ I1309A and β′ K1348A mutants showing the most detrimental defects in abortive synthesis on nifH and pspG promoters (Fig. 4A and C). The majority of the RNAP mutants seem to follow the effects of DksA on WT RNAP on three out of four promoters tested (with the exception of the pspA promoter). Thus, we propose that, in general, the BH and SW mutations do not override the impacts of DksA on σ54-dependent promoters. The observed effects on nifH, pspG and hrpL likely rely on intrinsic properties of each promoter sequence. Interestingly, both the pspA and pspG promoters share identical − 12 and − 24 elements and the transcription start site nucleotide (Fig. 4B and C, diagrams), yet they were activated by DksA to different extents (Fig. 4B and C). We propose that the sequence from − 10 region to − 1 region that constitutes the transcription bubble might be a major factor involved in fine-tuning DksA regulation via the BH and SW regions (see below).
Fig. 4.
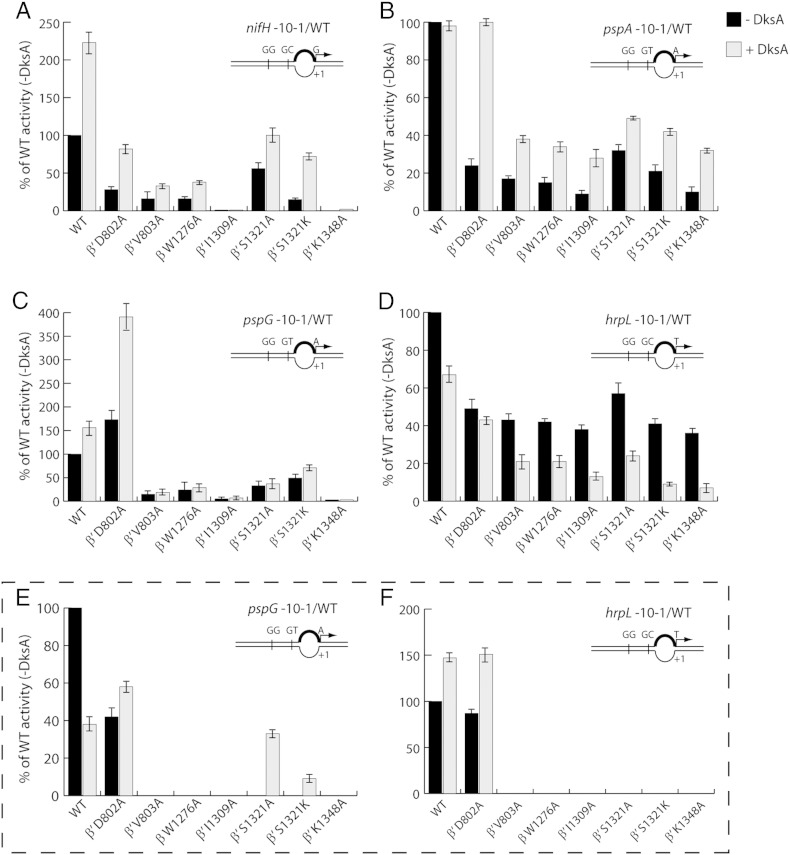
DksA shows kinetically diverse roles on σ54-dependent abortive synthesis. (A–D) Abortive synthesis was measured on four pre-melted σ54-dependent promoters with specific dinucleotide primers and radiolabelled nucleotides. The activity of each RNAP mutant in the absence and presence of DksA (black and grey bars, respectively) was expressed as a percentage of that of WT RNAP without DksA for each promoter. The full-length σ54 was used for abortive transcription with PspF1 -275 and dATP. (e and f) Abortive synthesis was measured on pspG and hrpL promoters with σ54ΔRI. Each experiment was reproduced in duplicate. The error bars represent standard deviation.
DksA works in synergy with σ54RI to impact on abortive synthesis
During DNA melting and loading, σ54 provides extensive interactions at the transcription bubble from − 10 to − 1 [46]. These interactions are probably mediated by σ54 Region I (residues 1–56). The σ54 Region I forms inhibitory interactions at the − 12 site prior to transcription activation and later couples the ATP hydrolysis energy to DNA melting. The σ54 Region II (residues 57–107) is functionally dispensable as it is absent in many σ54 species [47]. The σ54 Region III (residues 108–477) contains a winged helix–turn–helix domain that predominantly binds to the − 24 element [48]. We suspect that σ54RI may interact with the transcription bubble and the BH and SW network in the active centre either directly or indirectly and that it may play a role in forming RPO and abortive synthesis. To test this hypothesis, we generated σ54 with Region I deleted (σ54ΔRI) and performed abortive transcription assays on pspG − 10−1/WT and hrpL − 10−1/WT promoters (Fig. 4E and F). Deletion of σ54RI drastically reduced the ability of most BH and SW mutants to form abortive products on both promoters (compare Fig. 4E and F with Fig. 4C and D), suggesting that σ54RI indeed contributes to the BH and SW network. The presence of DksA seemed to partially rescue the abortive synthesis with β′ S1321A and β′ S1321K mutants (Fig. 4E), which may suggest that DksA adds another layer of interactions (direct or indirect) with the σ54RI-BH-SW network. Most intriguingly, deletion of σ54RI enables DksA to switch roles from stimulation to inhibition on the pspG promoter with the WT RNAP (compare Fig. 4C with 4E) and from inhibition to stimulation on the hrpL promoter with the WT and β′ D802A RNAPs (compare Fig. 4D with 4F). These observations argue that the influence of DksA relies not only on the intrinsic properties of the σ54-dependent promoter itself but also on the integrity of σ54RI.
BH and SW mutants are prone to backtracking and exhibit accelerated transcript-assisted hydrolysis
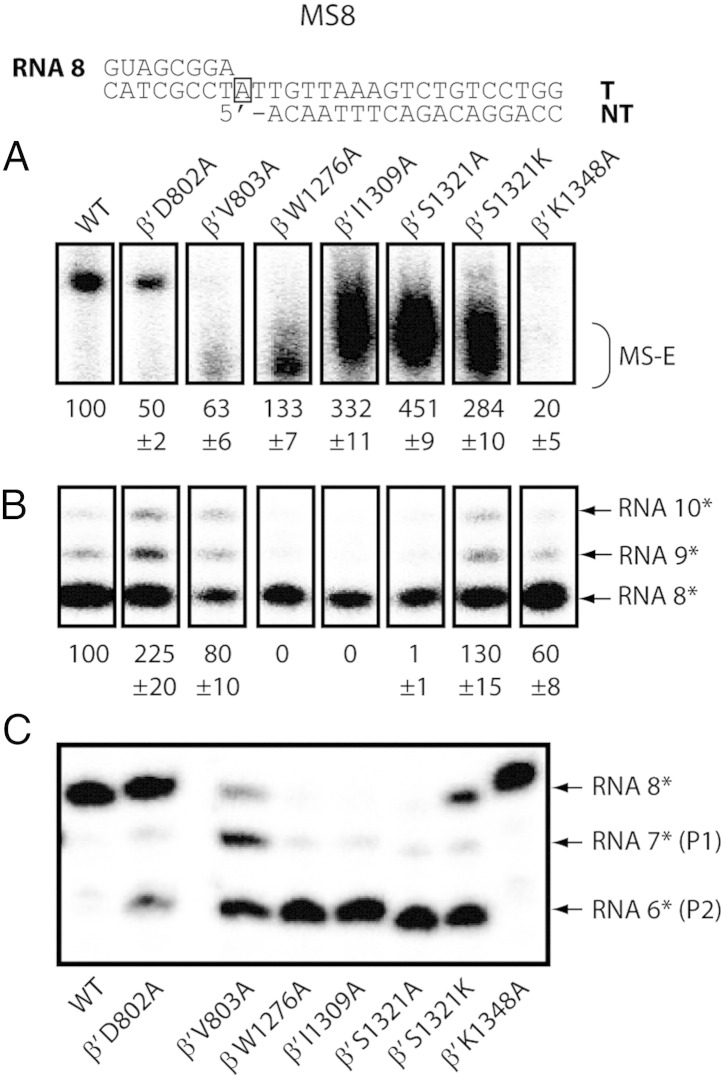
After assessing the impacts of BH and SW mutants on RPO formation and abortive synthesis, we next examined their abilities to elongate on a σ-independent DNA/RNA minimal scaffold [49]. The minimal scaffold is assembled with an 18-bp downstream DNA duplex and an 8-bp upstream RNA/DNA hybrid (MS8; Fig. 5). A gap was introduced after the “i + 1” site to accommodate the kinked DNA path [13]. An alternative minimal scaffold that harbours a 3′-CMP mismatch on the RNA was constructed for studying transcript-assisted hydrolysis [MS8(C); Fig. 5].
Fig. 5.
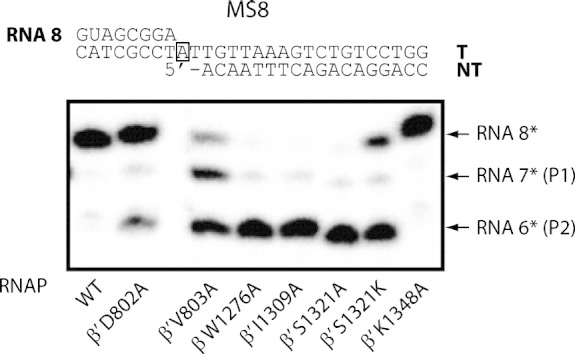
The BH and SW mutants are prone to backtracking and intrinsic RNA hydrolysis. The majority of the RNAP mutants can bind stably to the MS8 minimal scaffold (A) and transcribe with defects in the elongation state (B). The “i + 1” site is boxed on the template DNA strand. It should be noted that transcript-assisted cleavage takes place as soon as the RNAP is added to the MS8 scaffold. Hence, the RNA 8 is at different levels at the bottom of the gel. (C) Transcript-assisted cleavage was assessed on MS8 scaffold with RNA radiolabelled at the 5′-end. The cleavage of the ultimate and penultimate phosphodiester bonds are referred to as the P1 and P2 cleavage, respectively.
We first tested the elongation complex formation on a native gel (in the absence of σ factors). As seen in Fig. 5A, the majority of mutant RNAPs showed moderate (50–90% WT activity) to better-than-WT activities in the elongation complex formation. The β′ K1348A mutant showed the most detrimental defect (20% WT activity; Fig. 5A). Interestingly, many mutants showed different migration patterns to that of the WT RNAP, possibly due to differential clamp movements. To test whether the RNAP mutants could polymerise efficiently, we added a complementary nucleotide (cold UTP) to the 3′-end of the radiolabelled RNA 8. As seen in Fig. 5B, many of the tested RNAP mutants showed clear defects (below 50% WT activity) in transcript extension; such defects could not be entirely accounted for by weakened complex formation (compare Fig. 5B with 5A). In addition, many RNAP mutants generated RNA 10 through mis-incorporation (Fig. 5B).
To test whether these polymerisation-defective RNAP mutants favour backtracking and/or intrinsic hydrolysis, we performed transcript-assisted cleavage assays. Transcript-assisted cleavage occurs in a 1-bp backtracked state from the pre-translocated state. As the cleavage rate on fully matched 3′-NMP is extremely low, acceleration is usually achieved by mismatching the 3′-NMP [20], [50]. In the 1-bp backtracked state, the mismatched 3′-NMP occupies the E site (a non-base-pairing binding site) and assists the penultimate phosphodiester bond cleavage (P2 cleavage), possibly by enhancing Mg2 +II capture or the water molecule attack [20]. In the absence of a 3′-NMP (on the MS8 scaffold), the mutants β′ D802A, β W1276A, β′ I1309A, β′ S1321A and β′ 1321K all favoured P2 cleavage (Fig. 5C). The β′ V803A mutant exhibited a roughly equal proportion of P1 and P2 cleavage (Fig. 5C). It is likely that this RNAP mutant oscillates between a backtracked (P2 cleavage) and a pre-translocated state (P1 cleavage). Interestingly, mutants that show enhanced transcript-assisted cleavage are generally polymerisation defective (compare Fig. 5C with 5B). In a time-course cleavage experiment, we found that the P2 cleavage on MS8 was not a result of an observable prior P1 cleavage (Fig. S3). The inorganic pyrophosphate is thought to coordinate Mg2 +II and promote hydrolysis of the 3′-NMP occupying the “i + 1” site [17], [51]. After adding 0.5 mM PPi to the MS8 scaffold complex, we did not observe any alteration in the cleavage pattern (compare Fig. S4 with Fig. 5C), showing that the cleavage is not PPi dependent.
We next tested if these backtracked complexes induced by BH and SW mutations could re-initialise by NTP addition. Re-initiation of elongation was confirmed with WT and β′ D802A RNAPs only on the MS8 scaffold (Fig. S5). This observation suggests that not all backtracked states of the RNAP mutants are functionally equivalent, as some can be reversed back in to elongation whilst others were driven towards further cleavage.
Discussion
The differences in sequence between σ70 and σ54 are reflected in the different mechanisms of RPO formation at play for these two transcription paradigms [34], [35]. Although it is difficult to define precisely the relative location of the three σ54 domains within holoenzymes based on existing structural knowledge of σ70, a cryo-electron microscopy contour of the Eσ54–PspF complex suggests that σ54RI blocks access to the DNA loading channel, somewhat functionally reminiscent to the “place-holder” function of σ701.1 [52]. Here, we report an additional function of σ54RI through its interactions with the BH and SW regions (and possibly also DksA). σ54RI strongly stabilises the transcription bubble near the + 1 site on both strands, and RPO formed without σ54RI is unstable [45], [52]. A co-localisation of σ54RI with the BH/TL/SW at the + 1 site could explain why deletion of σ54RI has a detrimental impact on RPO formation and abortive synthesis with the mutant RNAPs (Fig. 2A and B). Although both σ701.1 and σ54RI may interact with the DNA loading channel at some point along the pathway from RPC to RPO, their precise local interactions with RNAP and their physical routes of displacement for or after DNA loading may well be different. This could explain why transcription activation of σ54 and of σ70 are differentially impacted by BH and SW mutations (this study) and why σ54 is not as sensitive to the T7 Gp2 protein that disrupts the normal displacement of σ701.1 [53], [54].
Three mechanistic models for DksA regulation on alternative σ-dependent transcription have been proposed [55]: (i) DksA potentiates alternative σ factors for their competitiveness, (ii) intrinsic properties of promoter DNA lead to different impacts by DksA and/or (iii) the availability of RNAP increases when DksA regulates σ70-dependent promoters (the passive model). Experiments carried out by Bernado et al. favour the passive model, where the authors did not observe any increased competitiveness for core RNAP of σ54 or transcription stimulation by DksA in vitro [43]. In this study, we provide evidence that DksA has direct and kinetically diverse impacts on σ54 promoters at the stage of abortive synthesis. We demonstrated that DksA stimulated abortive synthesis on nifH, pspA and pspG promoters and inhibited abortive synthesis on the hrpL promoter (Fig. 4). Notably, the TraR protein (a DksA homologue) has been shown to stimulate extracytoplasmic stress genes as regulated by the alternative σE in the absence of the cofactor ppGpp [56]. It is possible that DksA alters the energetic state on σ54-regulated promoters prior to the progression to the abortive cycle [42], or it works in synergy with the bridge helix and trigger loop to affect abortive synthesis. However, these possibilities do not explain why deletion of σ54RI affects DksA's role on abortive synthesis on pspG and hrpL promoters (Fig. 4E and F). Hence, we propose that a tripartite network involving σ54RI, DksA and the BH and SW regions is at play for DksA's regulation on σ54-dependent transcription.
Transcript-assisted RNA cleavage proceeds in a 1-bp backtracked conformation with the mismatched 3′-NMP slipping into the E site to facilitate the penultimate phosphodiester bond cleavage [20]. Without supplementing a mismatched 3′-NMP (either in cis or in trans), the rate of reaction is extremely slow [20], [50]. In this study, we have identified BH and SW mutants that are extremely efficient at RNA hydrolysis even on full-matched RNA/DNA hybrids. The accelerated cleavage is comparable to that observed in Thermus aquaticus RNAP that is usually 1–2 orders of magnitude faster than the hydrolysis mechanism in E. coli [16], [50]. The comparative studies with a mismatched scaffold showed that the hydrolysis activities were enhanced with our RNAP mutants. One possibility is that the 3′-RNA binds in the wrong register with respect to the transcription start site in the transcription complexes formed with the mutant enzymes. We believe that the observed phenotype points to a novel function of the BH and SW regions as an anti-backtracking ratchet and an RNA hydrolysis regulator. Disruption of such functions is particularly interesting in the β′ V803A mutant, as the RNAP is trapped in a sliding state (with both P1 and P2 cleavage equally likely; Fig. 5C).
Materials and Methods
Protein construction and purification
E. coli rpoB and rpoC genes harbouring single substitutions were mutagenised in plasmids pIA458, pIA545 and pIA661 and were sub-cloned into pVS10 vectors [57]. The reconstituted RNAP mutants were over-expressed in NovaBlue (Novagen) and purified as previously described [14]. E. coli PspF1 -275 (the AAA+ domain, residues 1–275) and Klebsiella pneumoniae σ54 were over-expressed in BL21(DE3) and purified as previously described [36]. E. coli σ70 was over-expressed in BL21(DE3) and purified via a nickel affinity column followed by a heparin chelating step. E. coli DksA was over-expressed in BL21(DE3) and purified via a nickel affinity column. Proteins were stored in TGED buffer [20 mM Tris–HCl (pH 8.0), 50 mM NaCl, 1 mM DTT, 0.1 mM ethylenediaminetetraacetic acid and 5% glycerol] at − 80 °C.
DNA probes
Linear DNA probes (88 nt long) representing E. coli lacUV5, S. meliloti nifH, E. coli pspA, E. coli pspG and P. syringae hrpL promoters were synthesised by Sigma-Aldrich with the highest purity (> 98%). The template strands were 5′-32P labelled and annealed to the mismatched non-template strands for binding assays [36].
RPO formation and minimal scaffold binding assays
The heparin-resistant RPO formation was assessed by mixing 50 nM radiolabelled DNA, 100 nM holoenzyme (1:4 ratio of E to σ), 4 μM PspF1 -275 and 4 mM dATP (for σ54-dependent RPO only) at 37 °C for 15 min. The complexes were challenged with heparin at 37 °C for 5 min before loaded on a 4% native gel and quantified by Aida.
Minimal scaffold binding assays were conducted by mixing an equimolar amount of RNAP and radiolabelled minimal scaffold (100 nM) at 37 °C for 15 min. Stable elongation complexes were resolved on a 4% native gel and quantified by Aida.
In vitro abortive transcription assays
In vitro abortive assays were performed as previously described [36]. Holoenzymes (100 nM, 1:4 ratio of E to σ) were allowed to assemble on specific promoter DNA probes, followed by addition of initiating dinucleotide primers and radiolabelled NTP for 15 min at 37 °C. In σ54-dependent transcription, 4 μM PspF1 -275 and 4 mM dATP were added to initiate transcription. To examine the impact of transcription factor on RPO formation, we incubated 2 μM DksA with the holoenzyme and DNA for 5 min at 37 °C before extension starts. To ensure that RPO formation was compared in a single-round transcription initiation, we added 0.2 mg/ml heparin during elongation (10 min at 37 °C). Promoter- and σ-independent minimal scaffold transcription assays were conducted using 100 nM core RNAP on 50 nM MS8 scaffolds at 37 °C for 15 min [58]. The forward polymerisation was performed by assessing the incorporation of 32P-UTP into RNA on MS8.
Cleavage assays
To assess backtracking and intrinsic cleavage of RNAP mutants, we radiolabelled the MS8 and MS8(C) minimal scaffolds on RNA 5′-ends. Intrinsic cleavage of RNA was allowed to proceed for 15 min at 37 °C before quenched. To test the pyrophosphorolytic activities of the RNAP mutants, we added 0.5 mM PPi to the reaction mixture as previously described [14]. To re-initiate elongation on backtracked complexes, we added 1 mM NTP mixture (ATP, GTP, CTP, UTP) to each cleavage reaction and incubated it 60 min to allow for transcript extension at 37 °C.
Acknowledgements
We thank Prof. S. Wigneshweraraj and Dr. M. Jovanovic for providing the E. coli rpoC 397 strain. We thank Prof. Nikolay Zenkin for his stimulating comments. This work was supported by Biological Sciences Research Council project grants (BB/J002828/1 and BB/G001278/1) to M.B.
Author Contributions: N.Z. and M.B. conceived and designed the experiments in the manuscript. N.Z., J.S., L.R. and A.S. performed the experiments. N.Z., J.S., X.D.Z., R.T., P.S. and M.B. interpreted data and wrote the paper.
Conflict of Interest: All authors declared no conflict of interest.
Edited by K. Severinov
Editor: Konstantin Severinov
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2015.09.005.
Contributor Information
Nan Zhang, Email: nan.zhang@imperial.ac.uk.
Martin Buck, Email: m.buck@imperial.ac.uk.
Appendix A. Supplementary data
Supplementary material
References
- 1.Ebright R.H. RNA polymerase: Structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 2.Lane W.J., Darst S.A. Molecular evolution of multisubunit RNA polymerases: Structural analysis. J. Mol. Biol. 2010;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane W.J., Darst S.A. Molecular evolution of multisubunit RNA polymerases: Sequence analysis. J. Mol. Biol. 2010;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruber T.M., Gross C.A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 5.Murakami K.S., Darst S.A. Bacterial RNA polymerases: The wholo story. Curr. Opin. Struct. Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 6.Landick R. RNA polymerase clamps down. Cell. 2001;105:567–570. doi: 10.1016/s0092-8674(01)00381-6. [DOI] [PubMed] [Google Scholar]
- 7.Chakraborty A., Wang D., Ebright Y.W., Korlann Y., Kortkhonjia E., Kim T., Chowdhury S., Wigneshweraraj S., Irschik H., Jansen R., Nixon B.T., Knight J., Weiss S., Ebright R.H. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gnatt A.L., Cramer P., Fu J., Bushnell D.A., Kornberg R.D. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 9.Mukhopadhyay J., Das K., Ismail S., Koppstein D., Jang M., Hudson B., Sarafianos S., Tuske S., Patel J., Jansen R., Irschik H., Arnold E., Ebright R.H. The RNA polymerase “switch region” is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava A., Talaue M., Liu S., Degen D., Ebright R.Y., Sineva E., Chakraborty A., Druzhinin S.Y., Chatterjee S., Mukhopadhyay J., Ebright Y.W., Zozula A., Shen J., Sengupta S., Niedfeldt R.R., Xin C., Kaneko T., Irschik H., Jansen R., Donadio S., Connell N., Ebright R.H. New target for inhibition of bacterial RNA polymerase: “Switch region”. Curr. Opin. Microbiol. 2011;14:532–543. doi: 10.1016/j.mib.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho M.X., Hudson B.P., Das K., Arnold E., Ebright R.H. Structures of RNA polymerase-antibiotic complexes. Curr. Opin. Struct. Biol. 2009;19:715–723. doi: 10.1016/j.sbi.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kettenberger H., Armache K.J., Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 13.Vassylyev D.G., Vassylyeva M.N., Perederina A., Tahirov T.H., Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 14.Wiesler S.C., Burrows P.C., Buck M. A dual switch controls bacterial enhancer-dependent transcription. Nucleic Acids Res. 2012;40:10878–10892. doi: 10.1093/nar/gks844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G., Campbell E.A., Minakhin L., Richter C., Severinov K., Darst S.A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J., Palangat M., Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat. Struct. Mol. Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nudler E. RNA polymerase active center: The molecular engine of transcription. Annu. Rev. Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozovskaya T.A., Rechinsky V.O., Bibilashvili R.S., Karpeisky M., Tarusova N.B., Khomutov R.M., Dixon H.B. The mechanism of pyrophosphorolysis of RNA by RNA polymerase. Endowment of RNA polymerase with artificial exonuclease activity. Biochem. J. 1984;224:645–650. doi: 10.1042/bj2240645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekine S., Tagami S., Yokoyama S. Structural basis of transcription by bacterial and eukaryotic RNA polymerases. Curr. Opin. Struct. Biol. 2012;22:110–118. doi: 10.1016/j.sbi.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Zenkin N., Yuzenkova Y., Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 21.Laptenko O., Kim S.S., Lee J., Starodubtseva M., Cava F., Berenguer J., Kong X.P., Borukhov S. pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J. 2006;25:2131–2141. doi: 10.1038/sj.emboj.7601094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lennon C.W., Ross W., Martin-Tumasz S., Toulokhonov I., Vrentas C.E., Rutherford S.T., Lee J.H., Butcher S.E., Gourse R.L. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 2012;26:2634–2646. doi: 10.1101/gad.204693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perederina A., Svetlov V., Vassylyeva M.N., Tahirov T.H., Yokoyama S., Artsimovitch I., Vassylyev D.G. Regulation through the secondary channel—Structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 24.Zenkin N. Multiple personalities of the RNA polymerase active centre. Microbiology. 2014;160:1316–1320. doi: 10.1099/mic.0.079020-0. [DOI] [PubMed] [Google Scholar]
- 25.Lee J.H., Lennon C.W., Ross W., Gourse R.L. Role of the coiled-coil tip of Escherichia coli DksA in promoter control. J. Mol. Biol. 2012;416:503–517. doi: 10.1016/j.jmb.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutherford S.T., Villers C.L., Lee J.H., Ross W., Gourse R.L. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendrickson E.L., Plotnikova J., Mahajan-Miklos S., Rahme L.G., Ausubel F.M. Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J. Bacteriol. 2001;183:7126–7134. doi: 10.1128/JB.183.24.7126-7134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heurlier K., Denervaud V., Pessi G., Reimmann C., Haas D. Negative control of quorum sensing by RpoN (sigma54) in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2003;185:2227–2235. doi: 10.1128/JB.185.7.2227-2235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joly N., Engl C., Jovanovic G., Huvet M., Toni T., Sheng X., Stumpf M.P., Buck M. Managing membrane stress: The phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 2010;34:797–827. doi: 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 30.Studholme D.J., Buck M. The biology of enhancer-dependent transcriptional regulation in bacteria: Insights from genome sequences. FEMS Microbiol. Lett. 2000;186:1–9. doi: 10.1111/j.1574-6968.2000.tb09074.x. [DOI] [PubMed] [Google Scholar]
- 31.Wolfe A.J., Millikan D.S., Campbell J.M., Visick K.L. Vibrio fischeri sigma54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 2004;70:2520–2524. doi: 10.1128/AEM.70.4.2520-2524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buck M., Gallegos M.T., Studholme D.J., Guo Y., Gralla J.D. The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J. Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasse-Dwight S., Gralla J.D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc. Natl. Acad. Sci. U. S. A. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman L.J., Gelles J. Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation. Cell. 2012;148:679–689. doi: 10.1016/j.cell.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma A., Leach R.N., Gell C., Zhang N., Burrows P.C., Shepherd D.A., Wigneshweraraj S., Smith D.A., Zhang X., Buck M., Stockley P.G., Tuma R. Domain movements of the enhancer-dependent sigma factor drive DNA delivery into the RNA polymerase active site: Insights from single molecule studies. Nucleic Acids Res. 2014;42:5177–5190. doi: 10.1093/nar/gku146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang N., Joly N., Buck M. A common feature from different subunits of a homomeric AAA + protein contacts three spatially distinct transcription elements. Nucleic Acids Res. 2012;40:9139–9152. doi: 10.1093/nar/gks661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burrows P.C., Joly N., Buck M. A prehydrolysis state of an AAA + ATPase supports transcription activation of an enhancer-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9376–9381. doi: 10.1073/pnas.1001188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaney M., Grande R., Wigneshweraraj S.R., Cannon W., Casaz P., Gallegos M.T., Schumacher J., Jones S., Elderkin S., Dago A.E., Morett E., Buck M. Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: Insights into activator mechanochemical action. Genes Dev. 2001;15:2282–2294. doi: 10.1101/gad.205501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joly N., Rappas M., Buck M., Zhang X. Trapping of a transcription complex using a new nucleotide analogue: AMP aluminium fluoride. J. Mol. Biol. 2008;375:1206–1211. doi: 10.1016/j.jmb.2007.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang N., Buck M. Formation of MgF3−-dependent complexes between an AAA+ ATPase and sigma54. FEBS Open Bio. 2012;2:89–92. doi: 10.1016/j.fob.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magnusson L.U., Gummesson B., Joksimovic P., Farewell A., Nystrom T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul B.J., Berkmen M.B., Gourse R.L. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernardo L.M., Johansson L.U., Solera D., Skarfstad E., Shingler V. The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of sigma-dependent transcription. Mol. Microbiol. 2006;60:749–764. doi: 10.1111/j.1365-2958.2006.05129.x. [DOI] [PubMed] [Google Scholar]
- 44.Lloyd L.J., Jones S.E., Jovanovic G., Gyaneshwar P., Rolfe M.D., Thompson A., Hinton J.C., Buck M. Identification of a new member of the phage shock protein response in Escherichia coli, the phage shock protein G (PspG) J. Biol. Chem. 2004;279:55707–55714. doi: 10.1074/jbc.M408994200. [DOI] [PubMed] [Google Scholar]
- 45.Hutcheson S.W., Bretz J., Sussan T., Jin S., Pak K. Enhancer-binding proteins HrpR and HrpS interact to regulate hrp-encoded type III protein secretion in Pseudomonas syringae strains. J. Bacteriol. 2001;183:5589–5598. doi: 10.1128/JB.183.19.5589-5598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burrows P.C., Wigneshweraraj S.R., Buck M. Protein-DNA interactions that govern AAA + activator-dependent bacterial transcription initiation. J. Mol. Biol. 2008;375:43–58. doi: 10.1016/j.jmb.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 47.Cannon W., Chaney M., Buck M. Characterisation of holoenzyme lacking sigmaN regions I and II. Nucleic Acids Res. 1999;27:2478–2486. doi: 10.1093/nar/27.12.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doucleff M., Pelton J.G., Lee P.S., Nixon B.T., Wemmer D.E. Structural basis of DNA recognition by the alternative sigma-factor, sigma54. J. Mol. Biol. 2007;369:1070–1078. doi: 10.1016/j.jmb.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korzheva N., Mustaev A., Kozlov M., Malhotra A., Nikiforov V., Goldfarb A., Darst S.A. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 50.Yuzenkova Y., Zenkin N. Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10878–10883. doi: 10.1073/pnas.0914424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sosunov V., Sosunova E., Mustaev A., Bass I., Nikiforov V., Goldfarb A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bose D., Pape T., Burrows P.C., Rappas M., Wigneshweraraj S.R., Buck M., Zhang X. Organization of an activator-bound RNA polymerase holoenzyme. Mol. Cell. 2008;32:337–346. doi: 10.1016/j.molcel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bae B., Davis E., Brown D., Campbell E.A., Wigneshweraraj S., Darst S.A. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma70 domain 1.1. Proc. Natl. Acad. Sci. U. S. A. 2013;110:19772–19777. doi: 10.1073/pnas.1314576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wigneshweraraj S.R., Burrows P.C., Nechaev S., Zenkin N., Severinov K., Buck M. Regulated communication between the upstream face of RNA polymerase and the beta′ subunit jaw domain. EMBO J. 2004;23:4264–4274. doi: 10.1038/sj.emboj.7600407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nystrom T. Growth versus maintenance: A trade-off dictated by RNA polymerase availability and sigma factor competition? Mol. Microbiol. 2004;54:855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- 56.Grace E.D., Gopalkrishnan S., Girard M.E., Blankschien M.D., Ross W., Gourse R.L., Herman C. Activation of the sigmaE-dependent stress pathway by conjugative TraR may anticipate conjugational stress. J. Bacteriol. 2015;197:924–931. doi: 10.1128/JB.02279-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belogurov G.A., Vassylyeva M.N., Svetlov V., Klyuyev S., Grishin N.V., Vassylyev D.G., Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jovanovic M., James E.H., Burrows P.C., Rego F.G., Buck M., Schumacher J. Regulation of the co-evolved HrpR and HrpS AAA + proteins required for Pseudomonas syringae pathogenicity. Nat. Commun. 2011;2:177. doi: 10.1038/ncomms1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material