Figure 4.
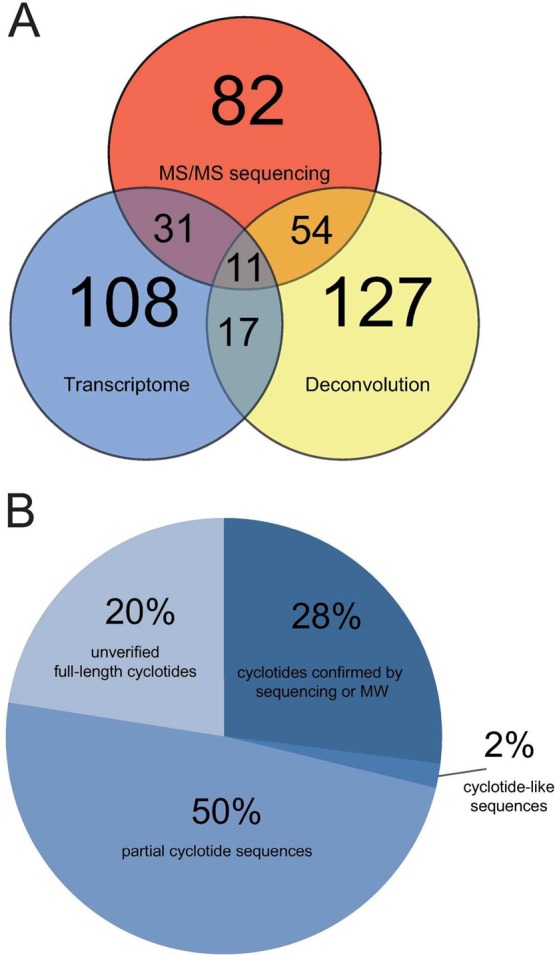
Comparative analysis of vitri cyclotides identified by transcriptome mining, mass deconvolution, and MS/MS sequencing. (A) An overview of vitri peptides discovered in this study is summarized by a Venn diagram. The number of identified cyclotides by each approach (transcriptome mining, mass deconvolution or MS/MS sequencing) is indicated by the numbers in each of the three interwheeling circles. The overlapping regions represent the numbers of identified cyclotides by combination of two or all three approaches. (B) A pie chart illustrates coverage for confirmation of predicted vitri cyclotides by MS. All predicted 108 vitri cyclotides identified by transcriptome mining were grouped in cohorts: (i) confirmed by sequence or molecular weight (28%), (ii) partial sequences from transcriptome mining (50%), (iii) cyclotide-like molecules (2%) and unverified full-length cyclotides (20%).
