Figure 3. On- and off-target indels in single K562 cells microinjected with HBB-targeting nucleases.
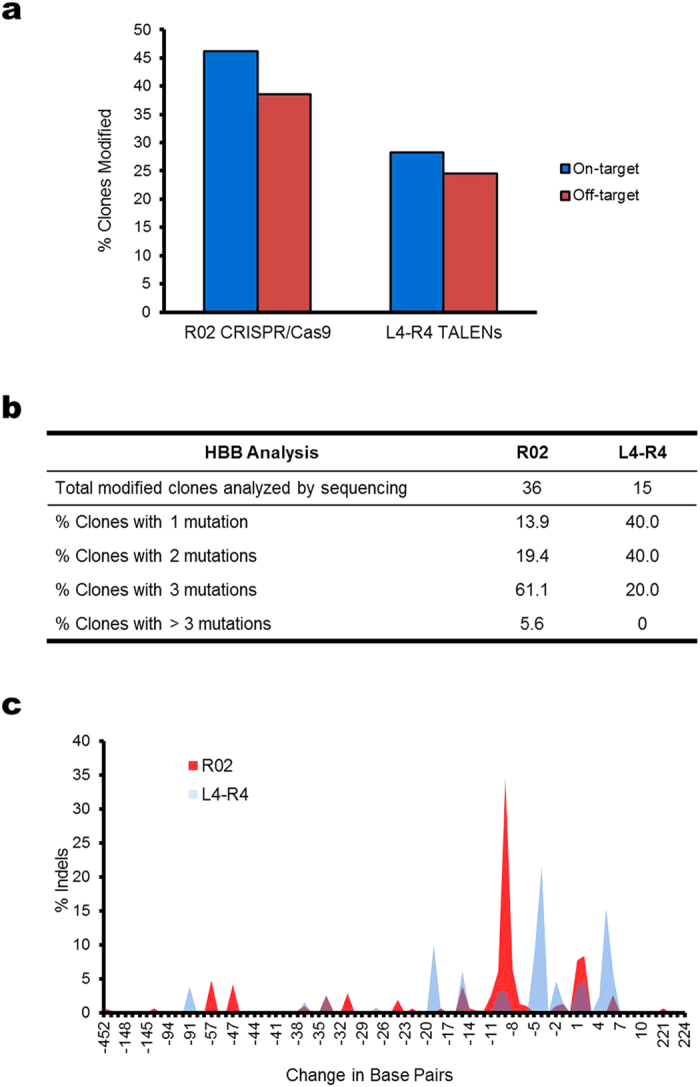
Cells microinjected with 200 ng/μL plasmids encoding L4-R4 TALENs or R02 CRISPR/Cas9 were expanded from single cells into clonal colonies in a 14 to16-day culture. T7E1 assay and Sanger sequencing were performed in individual clones and the number of clones having measurable indels was determined. (a) The percentages of clones with measurable on- and off-target indels. The off-target indels were detected in the GRIN3A locus and HBD for R02 CRISPR/Cas9 and L4-R4 TALENs respectively. N = 78 (R02) and 53 (L4-R4). (b) The percentage of clones having specific numbers of HBB mutations detected from 8–24 total sequencing reads. (c) Indel spectrum in cells microinjected with L4-R4 TALENs or R02 CRISPR/Cas9 determined using Sanger sequencing. The change in the number of base pairs resulting from NHEJ repair of DNA cleavage at the HBB target locus was compiled for each sequence read. The y-axis represents the percentage of indels with specified number of base pair changes.
