Fig. 1.
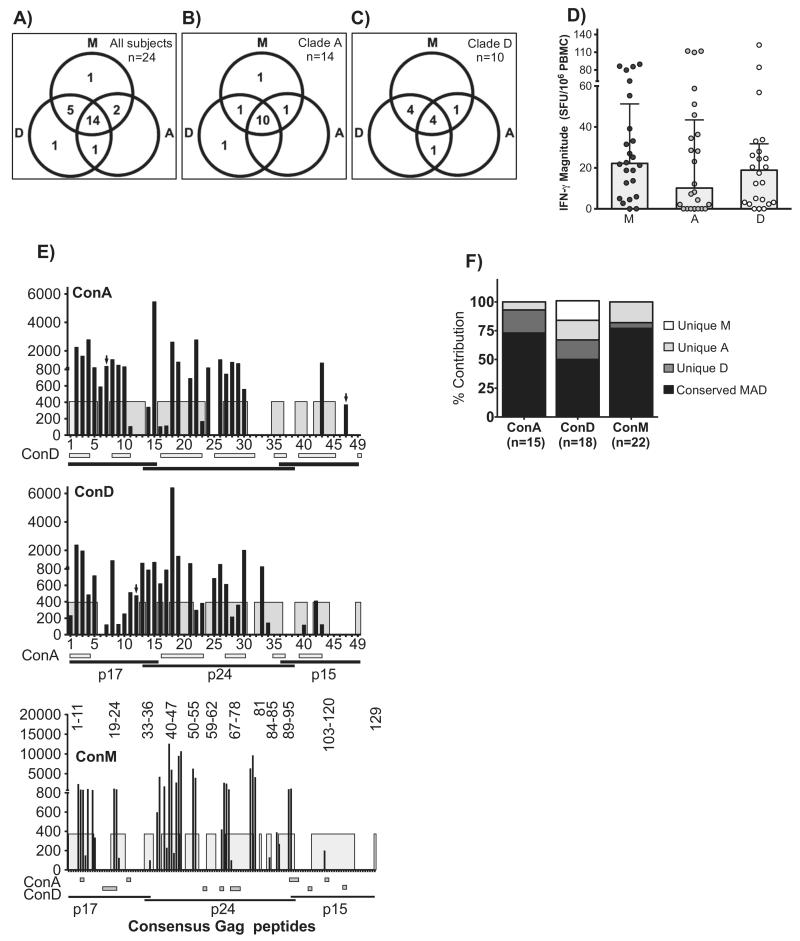
Gag-specific IFN-γ was detected in similar proportions of subjects across peptide sets, but ConM underestimated the breadths of response. The IFN-γ ELISpot assay was used to evaluate detection of Gag-specific IFN-γ response to ConM (n = 129), ConA (n = 49) and ConD peptides (n = 49) in 24 HIV clade A1 and D chronically infected subjects. Recognition of ConM, ConA and ConD peptides is illustrated in (A); and is subsequently stratified by clade A1- (B; n = 14) and D-infected subjects (C; n = 10). For each peptide set, total magnitudes were divided by the number of peptides evaluated to adjust for the varying numbers of peptides contained in each set. For example, magnitudes for the group M peptide set were divided by 129; and 49 divided magnitudes of the clade-based consensus peptides. (D) illustrates evaluations of adjusted IFN-γ magnitudes across peptide sets. Targeting individual ConA, ConD and ConM peptides by the 24 study subjects is chronologically illustrated in (E). Gray sections within the plot areas define regions with ≥90% sequence identity across sequences. The Y-axis indicates SFU/106 PBMCs while the X-axis displays the individual peptides. The ConM set has 129 peptides and cannot display on the X-axis; ConM peptide within the highly region across ConM/ConA/ConM are hence displayed as peptide numbers in the plot area. Horizontal bars below the X-axis define regions with ≥90% sequence identity between a given consensus clade peptide sequence and the plotted consensus peptide. Black arrows show peptides that uniquely detected virus-specific IFN-γ responses. Relative distribution of the detected IFN-γ breadth throughout unique and highly conserved regions across ConM, ConA and ConD is illustrated in (F).