Fig. 1.
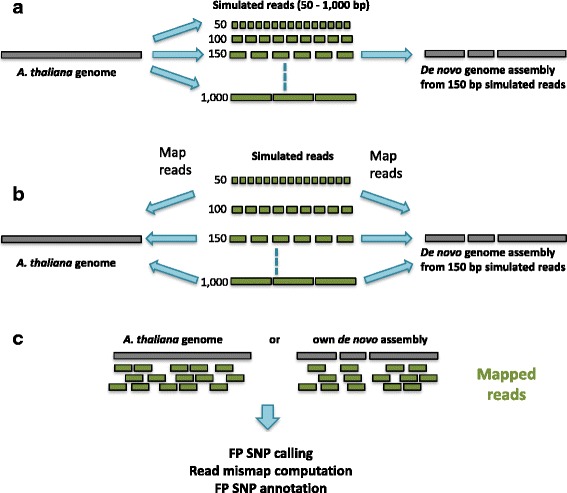
Experimental design. a The A. thaliana genome was used to generate simulated reads of different lengths. De novo assemblies were computed from the 150 bp read datasets using different assemblers. b With the assemblies as references, separate read mappings were carried out for each of the different read length datasets and with different combination of factor levels, using the original genome as a control. c SNP detection was carried out with different variant callers and the results were analysed to detect whether the mismatched reads causing the SNPs were due to mismapping. SNP annotation was performed to detect enrichment for particular genomic features at SNP positions
