FIGURE 3:
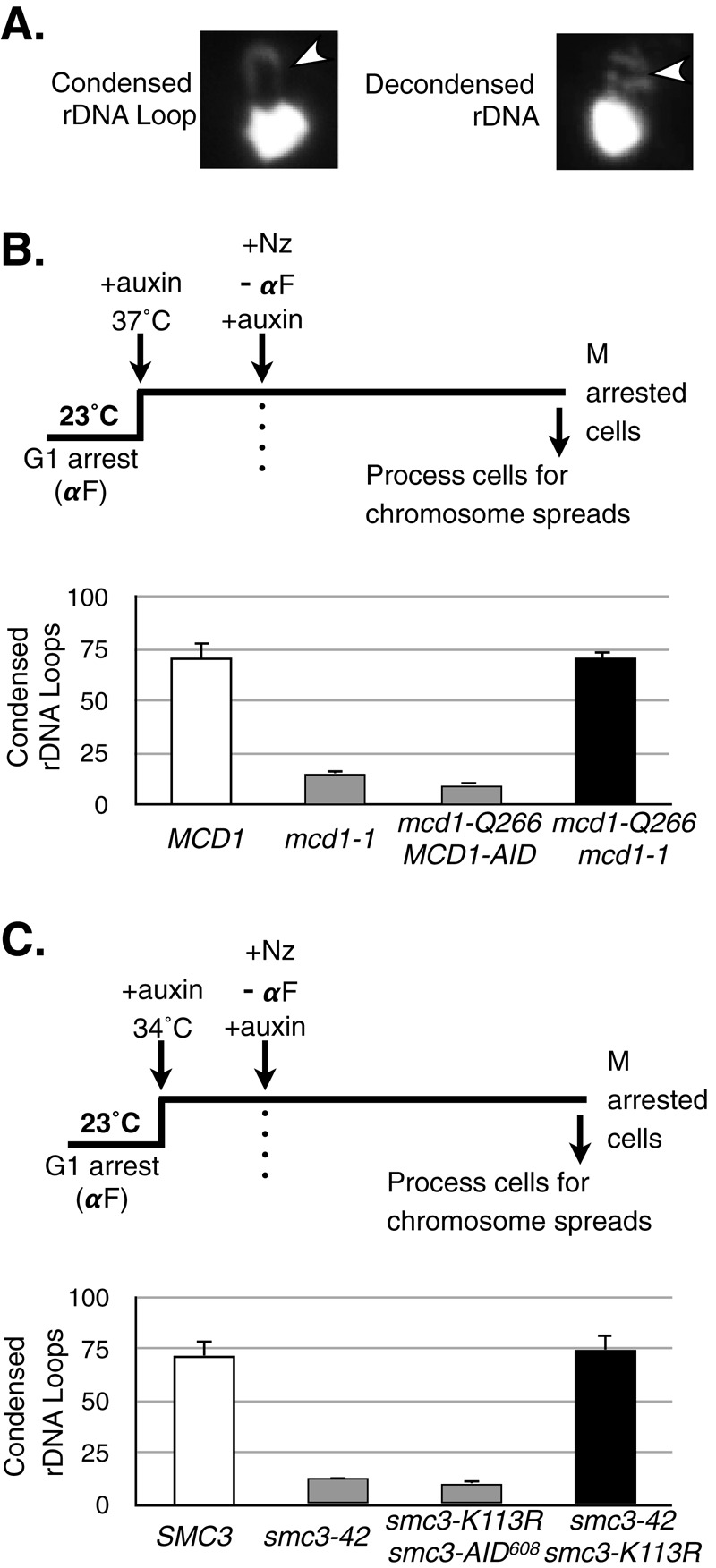
Analysis of chromosome condensation in MCD1 and SMC3 mutant allele strains. (A) Classification of rDNA morphology as assayed by chromosome spreads. Yeast spheroplasts were prepared, and chromosomes were spread on glass slides (see Materials and Methods). Chromosome masses were stained using DAPI and scored for morphology of the rDNA locus, which is adjacent to the bulk chromosome mass. A loop indicates a condensed rDNA locus (left), whereas a puff indicates a decondensed state (right), The rDNA is indicated by an arrowhead. (B) Haploid wild-type MCD1 (yVG3349-1B), mcd1-1 (yVG3312-7A), mcd1-Q266 MCD1-AID (yTE149), and mcd1-Q266-6MYC mcd1-1 (yTE42) strains were arrested in G1 phase at 23°C, auxin was added to degrade AID-tagged proteins, and shift to 37°C inactivated temperature-sensitive proteins. Cells were released from G1 at 37°C into medium containing auxin and nocodazole to arrest cells in mid M phase. Cells in mid M phase were processed for chromosome spreads and the rDNA morphology scored (Materials and Methods). The percentage of cells with condensed rDNA loops is plotted. At least 200 nuclei were scored for each genotype, and at least two biological replicates were completed for each genotype. (C) Haploid wild-type SMC3 (yTE45), smc3-42 (yVG3358-3B), smc3-K113R smc3-AID608 (yTE440), and smc3-42 smc3-K113R (yVG3473-1C) strains were synchronously arrested in mid M phase in auxin-containing medium as described in B, except that the temperature was up-shifted to 34°C. Cells were processed and scored for rDNA morphology and the data plotted as the percentage of cells with condensed rDNA loops as described in B. At least 200 DAPI-stained masses were scored for each genotype, and at least two biological replicates were completed for each genotype.
