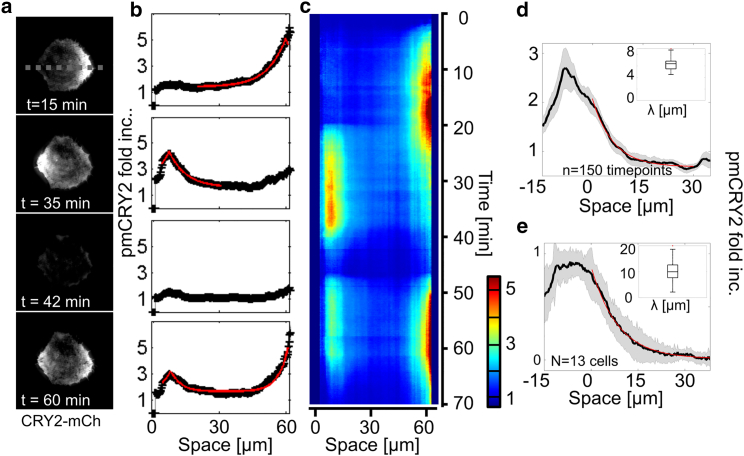
Figure 4.
pmCRY2 spatial distribution for a punctual activation. (a–c) CRY2-mCherry TIRF signals of a cell being successively activated in different points (a) and its corresponding kymograph (c) showing the quantified pmCRY2 profile along a horizontal line as exemplified for t = 15, 35, 42, and 60 min (b). First activations between time t = 2 min and t = 20 min in the right of the cell (one pulse every 25s). Second activation routine between t = 20 min and t = 35 min on the left of the cell. Third set of activations between t = 46 and 60 min on both sides. The decaying spatial distributions at steady state (black) were fitted with exponentially decreasing functions (red). The cell is initially plated on a 50-μm-diameter round pattern of fibronectin. (d) Quantification of the variability of the exponential gradient over time. An exponential gradient was established on one side of a round cell and maintained for >30 min (Movie S4). (Black line) Average gradient over 30 min; (gray shadow) 1 standard deviation. (Inset) Boxplot of the decay lengths measured for each time point (n = 150), λexp = 6.3 ± 1 μm. (e) Quantification of the variability of the exponential gradient for different cells. For each of the 13 cells, we used the time-averaged distribution of pmCRY2 to compute the average and SD of the pmCRY2 exponential gradient (see Materials and Methods for the normalization procedure). (Inset) Boxplot of the decay lengths measured for each cell, λexp = 10.5 ± 5 μm. To see this figure in color, go online.