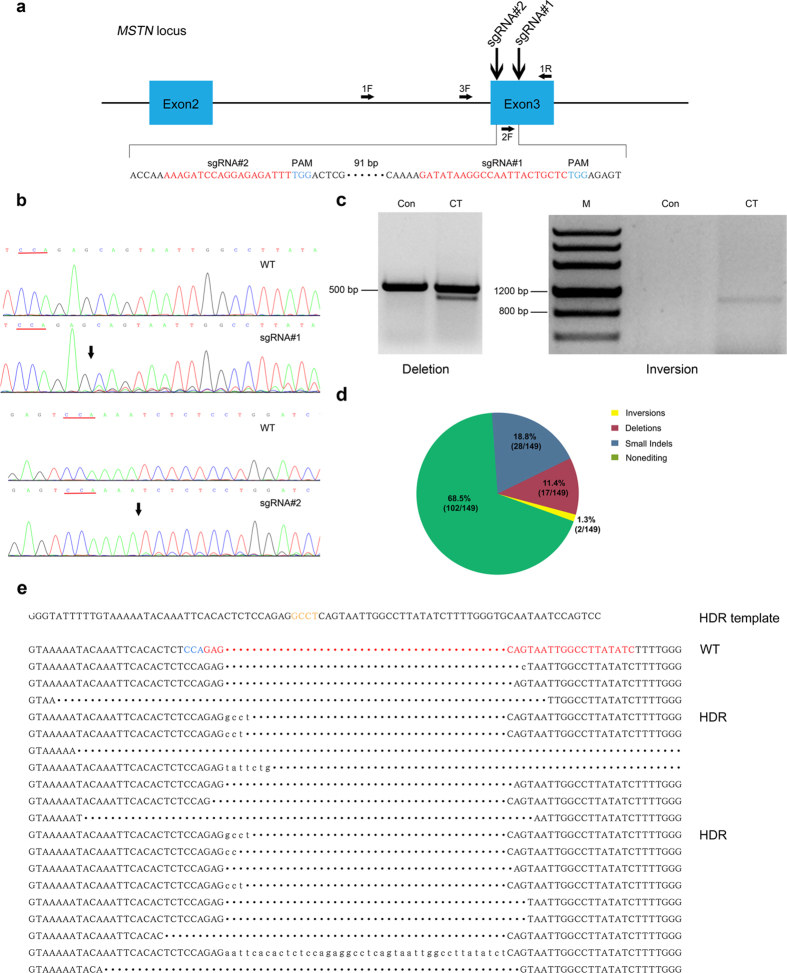
Figure 2. Editing of the porcine MSTN locus using the CRISPR/Cas9 system.
(a) Schematic representation of sgRNAs specific to exon 3 of the porcine MSTN locus. SgRNA targeting sequences are highlighted in red, and PAMs are highlighted in blue. (b) Genomic sequences of CRISPR target regions in wild-type PFFs and transfected PFFs, as indicated. PAMs are underlined in red and the cleavage sites are labeled with arrow. (c) Dual sgRNA-induced mutation at the porcine MSTN locus. PCR results for the targeted regions from co-transfected (CT) and un-transfected (Con) PFFs are shown. Both deletion and inversion bands were detected using specific primers. (d) The dual sgRNA-induced deletion and inversion frequency were further determined based on 149 amplicons covering the target sites. (e). Sequence of the ssDNA donor (upper panel). The newly added nucleotides are highlighted in orange. Sequencing of sgRNA#1 and the ssDNA donor induced mutations in the MSTN gene (lower panel). PCR amplicons spanning the target sites were analyzed for indels. The wild-type sequences are placed on the first line, while mutant TA clones are placed below. Target sites are indicated in red, PAM in blue, and inserted or substituted bases are presented in lower case. Clones showing correct HDR are labeled to the right of the sequences.