Table 3. 16S rRNA gene primer pair information and in silico amplicon statistics.
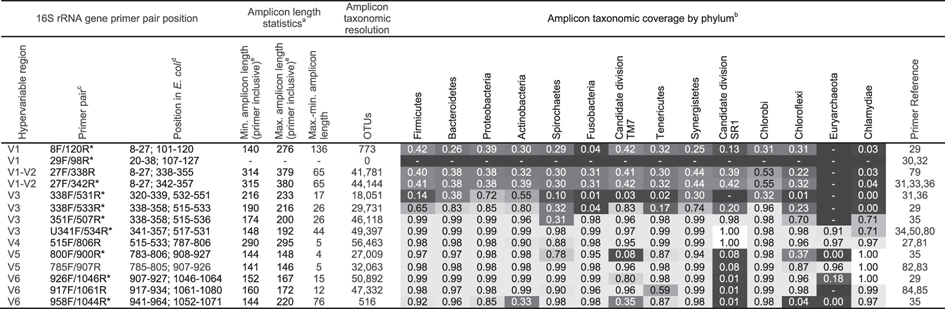
*Primer pair has been used in previous ancient microbiome studies. aLength statistics obtained from SILVA SSU 111 database. bIn silico estimate using PrimerProspector48 of the proportion of sequences in SILVA SSU 111 database (http://www.arb-silva.de) that the primer pair will amplify per phylum. Phyla are sequentially ordered from left to right by the number of species entries per phylum in the HOMD40. cPrimer pair naming conventions have changed through time and are not consistent across studies. dPosition of primer start and stop coordinates relative to Escherichia coli Genbank accession J01695. Full primer sequences are provided in Supplementary Table 2. eReported for 99% CI of the SILVA SSU 111 database. Outliers, which may include chimeras, are excluded. fOTUs generated per primer pair by de novo clustering of the Silva 111.1 database at 97% similarity. The total number of OTUs generated from full 16S rRNA gene sequences is 138,462.
