Abstract
Influenza virus delivers its genome to the host cytoplasm via a process of membrane fusion mediated by the viral hemagglutinin protein. Optimal fusion likely requires multiple hemagglutinin trimers, so the spatial distribution of hemagglutinin on the viral envelope may influence fusion mechanism. We have previously shown that moderate depletion of cholesterol from the influenza viral envelope accelerates fusion kinetics even though it decreases fusion efficiency, both in a reversible manner. Here, we use electron cryo-microscopy to measure how the hemagglutinin lateral density in the viral envelope changes with cholesterol extraction. We extract this information by measuring the radial distribution function of electron density in >4000 viral images per sample, assigning hemagglutinin density by comparing images with and without anti-HA Fab bound. On average, hemagglutinin trimers move closer together: we estimate that the typical trimer-trimer spacing reduces from 94 to 84 Å when ∼90% of cholesterol is removed from the viral membrane. Upon restoration of viral envelope cholesterol, this spacing once again expands. This finding can qualitatively explain the observed changes to fusion kinetics: contemporary models from single-virus microscopy are that fusion requires the engagement of several hemagglutinin trimers in close proximity. If removing cholesterol increases the lateral density of hemagglutinin, this should result in an increase in the rate of fusion.
Introduction
The influenza viral envelope contains three major viral proteins: hemagglutinin (HA), neuraminidase, and the M2 channel. Hemagglutinin and neuraminidase protrude from the viral surface in trimeric and tetrameric assemblies, respectively, forming spikes that are readily observable by electron microscopy (1, 2, 3, 4). Hemagglutinin is the sole protein component shown necessary and sufficient for viral membrane fusion with cellular target membranes (5, 6, 7), a key step in viral entry and subsequent infection of cells. This understanding in the field is that multiple hemagglutinin trimers contribute to optimal membrane fusion (8, 9, 10, 11). Therefore, spatial patterning and dynamics of hemagglutinin on the virion surface are expected to affect membrane fusion.
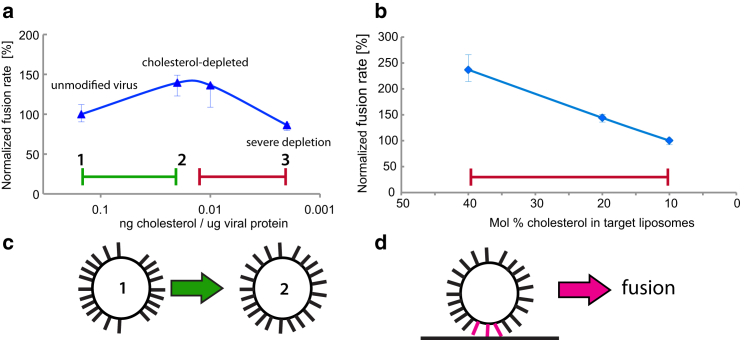
Influenza membrane fusion is sensitive to perturbation in either protein (12, 13, 14, 15, 16, 17, 18, 19) or lipid (20, 21, 22, 23, 24) components. Cholesterol has been a particular focus of lipid-based studies (20, 25), as the cholesterol content of the influenza envelope is greatly increased relative to the apical membrane of cells whence it buds (26). We have previously shown that extraction of cholesterol from the envelope of X-31 influenza virions produced a surprising result (Fig. 1): moderate extraction of cholesterol reduced fusion efficiency in a reversible manner but actually increased fusion rates, measured both by contents and by lipid mixing (27). Here, we use “fusion efficiency” to refer to the relative number of fusion events produced by a standardized quantity of virus, while rates are calculated from exponential fits to viral fusion kinetics. Severe extraction of cholesterol produced the expected behavior, reducing both fusion efficiency and fusion rates. By contrast, reducing cholesterol in target membranes reduced both fusion rate and efficiency as expected. This bimodal response profile to viral cholesterol levels clearly suggests multiple mechanistic roles for viral envelope cholesterol in membrane fusion.
Figure 1.
Bimodal response of influenza fusion rate to cholesterol extraction. Influenza fusion kinetics with target liposomes are plotted as cholesterol is extracted from the virus (a) or added to the liposomes (b). (Blue) Data are replotted from Domanska et al. (27) and show fusion of X-31 virus at different levels of cholesterol extraction with liposomes composed of 30:30:40 POPC/POPE/cholesterol (left plot) and unmodified X-31 virus with liposomes at different mole fractions cholesterol (right plot). These include a regime where fusion rates increase (green) with moderate cholesterol extraction from the virion (green) and one where they decrease with severe extraction from the virion or reduction of cholesterol in target liposomes (red). Schematized in (c) are the central findings of this article—that cholesterol extraction from the viral envelope (from point 1 to point 2 in a) causes a redistribution of hemagglutinin that reduces the average HA-HA spacing. This may explain the portion of the bimodal kinetic response where cholesterol extraction increases fusion rates (green region in a). Schematized in (d) is the theory that the activity of some number of HA trimers in close proximity is required for fusion. If this is true, then fusion rates should increase with increased HA spatial density. To see this figure in color, go online.
Here, we focus on the role of cholesterol in modifying hemagglutinin spatial distributions in the viral envelope and the consequences for influenza viral entry. Hemagglutinin has been observed to cluster in a cholesterol-dependent manner on the plasma membrane of transfected cells (28, 29), although recent imaging mass spectrometry data suggest that it may not colocalize with cholesterol in this context (30). However, it is not known how the spatial distribution of hemagglutinin responds to cholesterol in the more closely packed viral envelope, which has been suggested to bud in its entirety from specialized regions of the plasma membrane that may be enriched in cholesterol (31, 32, 33). The average cholesterol/phospholipid in X-31 virion samples is ∼1:1 (26, 27), and the glycoprotein (hemagglutinin or neuraminidase) spacing distance on the viral surface has been estimated at 110 Å from electron cryo-tomography (34), further supporting the idea that much of the viral surface may more closely resemble the clustered state of hemagglutinin in the plasma membrane of transfected cells than the unclustered state.
Studies in both transfected cells and model membranes have shown several potential roles for cholesterol in modulating influenza membrane fusion. These include stabilizing hemifusion intermediates or analogous transitions (25, 35, 36), altering membrane bending properties (37, 38), and changing the lateral distribution of membrane components (22, 28, 39). While transfected cells are a more accessible system for many biophysical studies, several lines of evidence suggest that the environment of the viral envelope may differ substantially from the cell surface in ways that affect hemagglutinin organization. Not only do influenza viral envelopes contain different amounts of lipid and cholesterol than the apical membranes of cells from which they bud (26), but even in pseudotyped viral systems (in which influenza hemagglutinin is a guest on another viral background), coexpressing the matrix protein from influenza increases the number of hemagglutinin molecules on the viral envelope.
Here, we have used electron cryo-microscopy to measure how the lateral distribution of hemagglutinin in the X-31 influenza viral envelope changes in response to cholesterol perturbation. We find that the HA-HA nearest-neighbor distance is reduced upon cholesterol extraction from the virion and increases again when cholesterol content is restored. These findings suggest an explanation for the observed bimodal trend in fusion rates: if cholesterol globally promotes fusion by lowering the free energy barrier to forming lipidic intermediates such as fusion stalks, this would explain a decrease in fusion rates when membrane cholesterol is reduced (red region in Fig. 1 a). However, if increased cholesterol also increases HA-HA average separation, this would decrease the rate at which multiple HA trimers are present at the fusion site and thus explain the increase in fusion rates at moderate cholesterol extraction from the viral membrane (green region in Fig. 1 a).
Materials and Methods
Influenza virus
Egg-grown influenza virus X:31 (H3N2 A/Aichi/68) was purchased from Charles River Laboratories (Wilmington, MA). Cholesterol was extracted by incubating viral samples at 0.5 mg/mL viral protein with 0, 5, 10, or 20 mM methyl-β-cyclodextrin (MβCD) at 37°C for 30 min. Virus was then reisolated from cyclodextrin-cholesterol complexes via centrifugation at 4°C, 14,000 rpm for 40 min, and resuspended in PBS (phosphate-buffered saline) (100 mM sodium phosphate and 150 mM NaCl, at pH 7.2). Unmodified control and cyclodextrin-treated virus samples were incubated identically; unmodified virus was used at 2 mg/mL viral protein, while MβCD-treated virus was used at 4 mg/mL of viral protein.
Tissue culture ID50 determination
MDCK cells were seeded at 7.5 × 104 cells/mL in a 96-well tissue-culture-coated plate and cultured overnight in DMEM media + 10% fetal bovine serum at 37 C, 5% CO2. Cells were washed and then incubated with DMEM + 0.1% fetal bovine serum containing the specified dilution of X-31 influenza virus. Infections were performed in replicas of four. Plates were incubated at 37 C, 5% CO2 for three days, and cytopathic effect was scored on each day. TCID50 values were calculated as the lowest dilution of virus with ≥50% cells/well infected. TCID50 values were obtained using both 10-fold and threefold dilutions and yielded the same ratio of values between viral samples.
Cholesterol replenishment of MβCD-treated influenza virus
Cholesterol-depleted influenza virus was treated with cholesterol/MβCD complex at 20 mM final concentration. Five milligrams of cholesterol were dissolved in 600 μL of chloroform, dried under nitrogen, and then placed under vacuum for 1 h. The resulting chloroform film was hydrated with 20 mM MβCD in PBS-citrate (10 mM phosphate, 90 mM citrate, and 150 mM NaCl, at pH 7.4) at a 1:1 cholesterol/MβCD molar ratio. The mixture was sonicated for 3 min and incubated in a water bath at 37°C overnight. The solution was filtered to remove cholesterol precipitate, and the ratio of remaining cholesterol to MβCD was estimated as 1:5.8. The virus was mixed with cholesterol/MβCD complex at a 2:1 ratio, and PBS-citrate was added to obtain a final concentration of 5 mM MβCD. This mixture was incubated at 37°C for 30 min, and then virus was isolated by centrifugation at 4°C and 14,000 rpm for 40 min followed by resuspension in PBS. The use of MβCD for cholesterol extraction and delivery and the separation of MβCD from membranes subsequent to treatment have been well-described previously (40, 41). Cholesterol concentrations were estimated using an Amplex Red cholesterol oxidase assay (Life Technologies, Grand Island, NY).
Labeling with anti-HA Fab fragment
Monoclonal anti-HA IgG HC3 was a gift from J. Skehel. Fab fragments were generated by IgG papain cleavage and purified according to the manufacturer’s protocol (Thermo Scientific, Rockford, IL). Virus was incubated with Fab HC3 fragments for 1 h at room temperature. The w/w ratio of viral protein to Fab was estimated at 4:1. Unbound Fab fragments were removed by centrifuging viral particles using Vivaspin500 concentrators (Vivaproducts, Littleton, MA) with a 100-kDa cutoff. Samples were centrifuged at 4°C, 12,000 g for ∼10 min.
Electron cryo-microscopy
Purified samples were vitrified as reported previously using a manual plunge-freezing device (42). Briefly, ∼3 μL of sample were applied to a glow-discharged, perforated carbon-coated grid (Cat. No. 2/2-4C C-flats; ProtoChips, Raleigh, NC), blotted with filter paper to near dryness, and rapidly plunged into liquid ethane. Fiducial gold particles were added to some samples to allow additional tomographic screening. The grids were stored in liquid nitrogen and then maintained in the Tecnai F20 Twin transmission electron microscope (FEI, Hillsboro, OR) at −180°C using a model No. 626 cryo-stage (Gatan, Pleasanton, CA). Low-electron dose images were recorded at a magnification of 29,000× operating at 120 kV, with a nominal defocus of −3 μm. All images were recorded with a 4K × 4K pixel charge-coupled device camera (Gatan), and the pixel size was 0.37 nm at the specimen level.
Radial distribution analysis
Square regions within each virion were manually selected from transmission electron micrographs using the EMAN software suite (43). Selected regions were 158 pixels or 585 Å in each dimension. In each micrograph examined, all virions where a distinct 585 × 585 Å region could be visualized were analyzed. Radially averaged Fourier transforms were computed for each of these regions (code freely available at https://github.com/kassonlab/em-spatial-analysis) in a procedure based on the one used by the EMAN CTFit program (44). Approximately 4000 such squares were selected and Fourier spectra computed per viral sample. The average of all Fourier spectra in a sample yielded the radial distribution function for electron density in the virion (transmission images of the viral interior include the projection of the top and bottom of the viral envelope as well as viral contents). Error analysis was performed via bootstrap resampling (1000 samples) of individual Fourier spectra.
Surface area analysis
Ellipses were manually fit to the viral perimeter in transmission electron microscopy images of 100 randomly selected virions in each sample. Surface areas were then calculated using the ellipse axes and approximating each virion as a prolate spheroid (or a sphere if the axes were equal). Viruses with ill-defined margins or ones too filamentous for accurate ellipse fitting were excluded from the surface-area analysis.
Results
We have used electron cryo-microscopy to measure the spatial distribution of hemagglutinin in the X-31 viral envelope and determine how this distribution shifts in response to cholesterol extraction and readdition. Because the contrast in images of unlabeled and unstained influenza virions did not permit robust automated classification of hemagglutinin and neuraminidase spikes in tomographic data, we developed an alternate approach. We measured radial distribution functions of electron density in transmission electron micrographs of influenza virions by computing radially averaged Fourier spectra. Hemagglutinin density was then assigned to particular peaks in these spectra by comparing spectra of samples with and without an anti-HA monoclonal antibody. This process yielded measurements of hemagglutinin lateral distribution in the viral envelope. Both the cholesterol manipulations and the process of measuring radial distribution functions are described in more detail below.
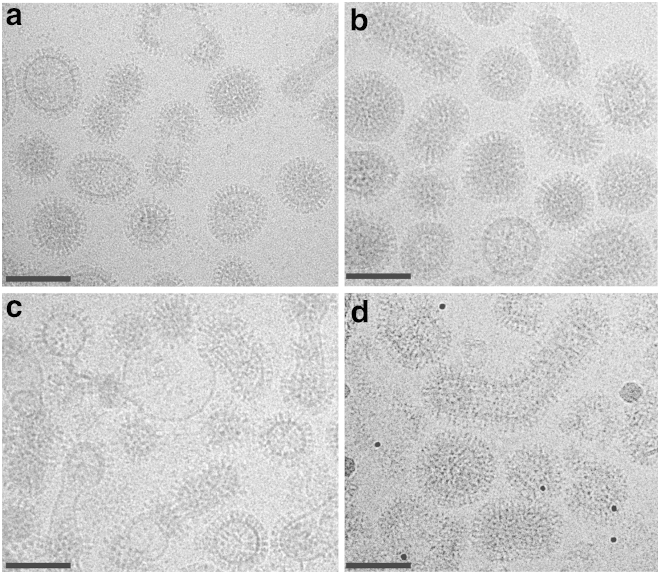
Cholesterol was manipulated in the viral envelope, using methyl-β-cyclodextrin as a reagent to extract or deliver sterol at the membrane (39, 45, 46). To match prior studies on fusion kinetics, egg-grown X-31 influenza virions were used. Incubation with 10 mM MβCD for 30 min yielded an 89 ± 3% reduction in viral envelope cholesterol, while incubation with 20 mM MβCD yielded at 97 ± 0.7% reduction on the viral lots used for electron microscopy, as measured via cholesterol oxidase assays. Isolation of the virus and further incubation with 20 mM MβCD that had been preloaded with cholesterol increased the cholesterol content of isolated virions to 144% of the native starting level. Unmodified X-31 viral samples were treated identically to those undergoing cholesterol extraction except for the absence of MβCD. The initial virus preparation contained some membrane particles that are not infectious virions and these were coisolated with virus throughout the procedure. If these particles were to absorb cholesterol from MβCD differently from infectious virus, this would introduce some error into the sterol quantitation. Tissue culture ID50 values for cholesterol-depleted virus were ∼16% of unmodified virus, and values for cholesterol-replenished virus were 100% of unmodified virus, showing full restoration of infectivity (see Materials and Methods for details). These three samples—unmodified virus, cholesterol-depleted, and cholesterol-replenished—were used for measurement of hemagglutinin spatial patterning via electron microscopy. Sample micrographs are shown in Fig. 2.
Figure 2.
Electron cryo-micrographs of X-31 influenza virions at different cholesterol levels. Unmodified virus is shown in (a), virus that has been cholesterol-depleted using 20-mM MβCD is shown in (b), virus that has been cholesterol-replenished is shown in (c), and cholesterol-depleted virus with bound HC3 Fab fragments is shown in (d). Images were obtained on a Tecnai F20 Twin transmission electron microscope (FEI) at 29,000× magnification. Scale bars = 100 nm.
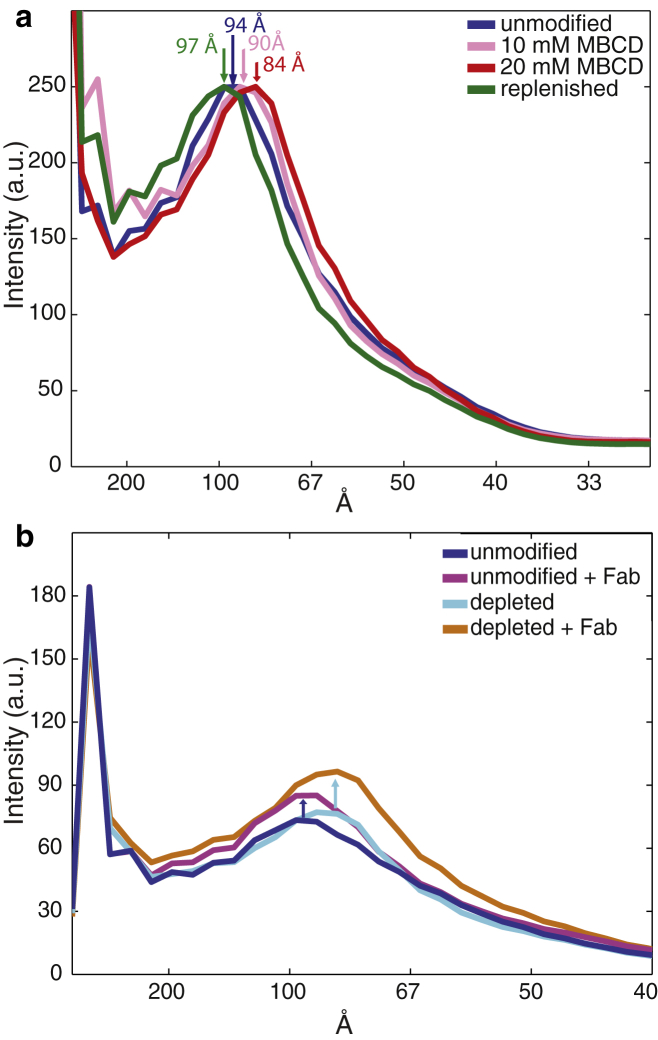
Hemagglutinin lateral distribution was measured as follows. For each sample, ∼4000 virions were identified, and square regions were selected in the interior of the virus on two-dimensional projection in transmission electron microscopy images. These regions of the image contain information on the top and bottom of the viral envelope as well as the interior contents. A radially averaged Fourier spectrum was computed for each image (see Materials and Methods for details and Fig. S1 in the Supporting Material for illustration), and these one-dimensional spectra were averaged to yield a single average radial distribution for the sample (Fig. 3). Error analysis was performed via 1000-fold bootstrap resampling. Samples at each cholesterol condition were also prepared with anti-HA Fab fragments bound, and radial distribution spectra were computed via an identical procedure. Fig. 3 b shows an overlay of spectra for unmodified virus with and without Fab. The increased density in the peak at 84–97 Å separation with Fab treatment allows assignment of the hemagglutinin density to that peak, although other viral components may make up a portion of the peak as well. This spacing corresponds roughly to the glycoprotein spike spacing value of 110 Å reported previously (34), particularly because this value lumps HA and neuraminidase spikes together. This distribution is also consistent with the distribution of glycoprotein spacing reported in a second tomographic study (47).
Figure 3.
Radial distribution functions showing hemagglutinin spatial distribution in response to cholesterol. (a) Average radial distributions of electron density plotted for unmodified virus, cholesterol-depleted virus with either 10 or 20 mM MβCD, and cholesterol-replenished virus. Lateral shifts in the peak at 84–97 Å show changes in hemagglutinin nearest-neighbor distances. (b) Average radial distribution functions of electron density plotted for unmodified virus and cholesterol-depleted virus with and without bound Fab fragments. The increase in density at the 94 Å peak allows assignment of hemagglutinin density to this peak. Confidence intervals of 95% computed via bootstrap resampling as well as confidence intervals for individual virions are plotted in Fig. S2.
Using this assignment of HA density, the shift in radial distribution spectra with cholesterol extraction can be interpreted in terms of changes to HA spatial distribution. Although the radial distribution function is computed from a cross section of the virus, the high electron density of HA relative to other viral components, and particularly the abundance of HA relative to other proteins in the viral envelope, makes this a reasonable approximation (48). In unmodified X-31 virus, the mode of the HA-HA distribution peak lies at 94 Å (Fig. 3 a), close to the value of 110 Å for the average glycoprotein spacing distance reported from hand-labeling of tomographic data (34). The mode of the HA-HA distribution shifts to 84 Å in cholesterol-depleted virus (2% cholesterol remaining) and shifts back to 97 Å in cholesterol-replenished virus (cholesterol 144% of unmodified virus). Intermediate cholesterol extraction using 10 mM instead of 20 mM MβCD results in an intermediate shift of the HA-HA distribution mode, to 89 Å. Accompanying error analyses are plotted in Fig. S2, and HA-HA distributions are plotted for unmodified virus versus cholesterol extraction with 5, 10, and 20 mM using a second viral lot in Fig. S3. The computed Fourier spectra primarily capture nearest-neighbor distance; higher-order neighbor distances overlap with other portions of the spectrum and are either not as regular (and thus would be not visible in the averaged spectra) or not resolvable. Together, these data indicate that the average HA-HA separation decreases upon cholesterol extraction from the X-31 influenza viral envelope and then expands again upon readdition of cholesterol.
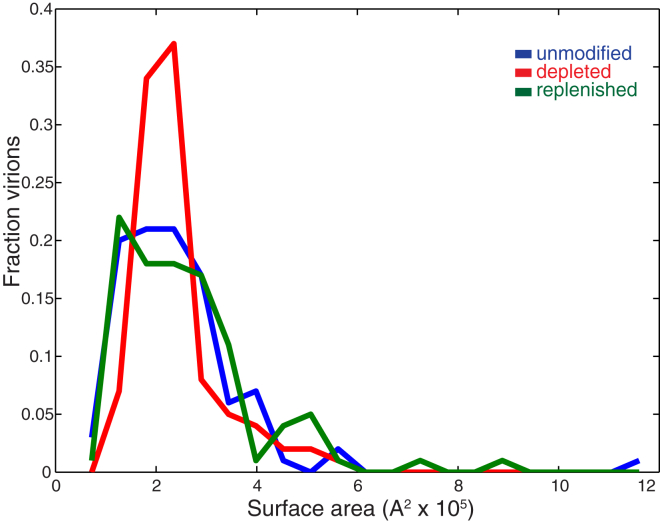
Shrinkage of overall viral surface area upon cholesterol extraction would provide a straightforward explanation for the change in hemagglutinin spacing, but cholesterol extraction from the viral envelope did not significantly change virion surface area. To estimate surface area, ellipses were manually fit to the viral envelope for 100 randomly selected virions in each sample, and the surface was modeled as a prolate spheroid. Surface area histograms for each sample are plotted in Fig. 4; there was no significant change in median surface area among unmodified, cholesterol-depleted, and cholesterol-replenished virions as measured by the Wilcoxon Rank Sum Test and no significant change in surface area distribution measured by the Kolmogorov-Smirnov test with a Bonferroni multiple hypothesis correction. If HA-HA spacing were uniform on the viral surface and the change observed upon cholesterol extraction were entirely due to surface area shrinkage, one would expect a 20% decrease in virion surface area; this was not observed. Similarly, there were no significant changes in the length/width ratio of virions (Fig. S4). These results are perhaps predictable, as cholesterol has a profound condensing effect on lipid bilayer surface area, where the magnitude depends on the chemical identity of the phospholipid tails (49, 50, 51).
Figure 4.
Virion surface area distributions at different levels of cholesterol extraction. Histograms of calculated viral surface area are plotted for unmodified, cholesterol-depleted, and cholesterol-replenished influenza virus. Median surface areas and 90% confidence intervals are 2.2 × 105 Å2 (1.1 × 105 to 4.4 × 105) for unmodified virus, 2.1 × 105 Å2 (1.5 × 105 to 4.1 × 105) for cholesterol-depleted virus, and 2.3 × 105 Å2 (1.2 × 105 to 5.0 × 105) for unmodified virus. Median surface areas and surface area distributions do not significantly differ among the three distributions (Wilcoxon Rank Sum test and Kolmogorov-Smirnov test with Bonferroni correction), although a statistically insignificant difference in the width of the main peak can be observed. To see this figure in color, go online.
Discussion
Here, we have measured the distance between hemagglutinin trimers on the influenza viral surface in response to cholesterol manipulation. Depletion of cholesterol from the viral envelope causes a decrease in typical HA-HA spacing that is reversible upon cholesterol replenishment. This closer spacing of hemagglutinin trimers may at first seem counterintuitive when compared to previous data showing cholesterol-dependent clustering of HA in the plasma membrane of transfected cells. However, given contemporary models for influenza budding and the high average sterol composition of influenza virus, it may be that the clustered environment in the plasma membrane represents a large fraction of the viral envelope.
We propose that the shift in hemagglutinin spacing is caused by lipid repartitioning between HA-containing and non-HA lipidic contexts. We propose a model based on three fundamental assumptions:
-
1)
The viral envelope is spatially heterogeneous and, in a hemagglutinin-centric view, can be described as near-HA or far-HA. Because HA is used as the frame of reference, HA movement is not considered explicitly.
-
2)
Lipids are in exchange between near-HA and far-HA portions of the viral envelope. Our model is thus a thermodynamic one in that it considers the average distributions of lipids and HA at equilibrium before and after cholesterol extraction.
-
3)
We propose that the chemical potential of cholesterol and lipids (the free energy of exchange between near-HA and far-HA portions) depends on the mole fraction cholesterol in the near-HA portion of the envelope, xchol_near. Our model does not specify the mole fraction cholesterol in the far-HA portion xchol_far, only that the chemical potential is insensitive to this mole fraction.
Consider near-HA and far-HA regions in exchange. One can express the free energy of each region as
| (1) |
where αij and βij are pairwise interaction energies between components i and j, γi is the interaction energy between component i and HA, and xi is the mole fraction of component i.
At equilibrium, no net flux between the near and far regions dictates that ∂Gnear/∂ni = ∂Gfar/∂ni for each exchangeable component i. Because we have postulated that the near-HA region is more sensitive to mole fraction cholesterol than the far-HA region, ∂Gnear/∂nchol-near > ∂Gnear/∂nchol-far. Cholesterol extraction reduces (nchol-near + nchol-far), so by the above relationships cholesterol will redistribute from the HA-far region to the HA-near region after extraction. We also propose that if the cholesterol extraction is substantial, other components will redistribute from the HA-near region to the HA-far region, thus increasing xchol-near to help compensate for the loss of cholesterol. If the total nchol-near does not increase from initial equilibrium to postextraction equilibrium, then the number of lipids Nnear will decrease. Because cholesterol can have a substantial condensing effect on area per lipid, a large decrease in xchol-near could mitigate any area reduction from decreasing Nnear; we hypothesize that the decrease in xchol-near is moderate due to postextraction repartitioning and that this combined with the decrease in ni-near for other phospholipids i causes an area reduction in the HA-near region, resulting in the observed decrease in HA-HA spacing.
Because many physical details of spatial organization on the virion surface remain undetermined, this model remains intentionally broad, and a number of different combinations of coefficients could satisfy the observed changes. There has been much work done on the theory of cholesterol-phospholipid interactions, particularly with regard to liquid-liquid phase coexistence (52, 53, 54, 55); our model draws inspiration from this prior work, and a number of these more specific models could explain our observations. However, we do not have sufficient data at this time to definitively support one model over another. Maintaining uncertainties regarding the identities of the exchanging components and the potential contribution of any stoichiometric complexes limits the specificity of our proposed model somewhat, but hopefully preserves its robustness as these details are uncovered.
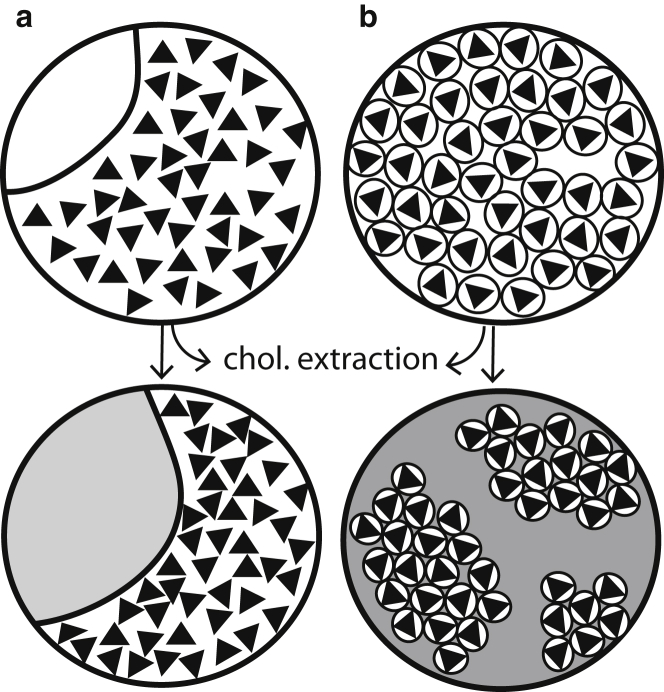
The consequences of our model for hemagglutinin spacing also depend on the size of the near-HA portions of the membrane. In one limit, the near-HA regions are large and contain many HA trimers (Fig. 5 a), similar to traditional nanodomain models of cholesterol-dependent HA clustering (28, 29, 56). However, these HA-containing regions likely comprise a majority of the viral envelope surface area because the HA:NA ratio has been measured at ∼10:1 (3, 34, 57). In this case, extraction of cholesterol will causes a decrease in the near-HA membrane surface area, and the HA-HA spacing will reduce as a consequence of an equal number of HA being distributed in a smaller region of the membrane (Fig. 1 c). In the opposite limit, each HA trimer is surrounded by a local lipid context (Fig. 5 b) of radius <5 nm (limited by the HA-HA spacing of ∼94 Å in unmodified virus). This is similar to a lipid shell model or annular lipid model (58, 59). These single-trimer nanodomains may tend to cluster, driven by line tension along the border between HA-containing and non-HA-containing regions of the membrane. Cholesterol extraction from the envelope will once again cause repartitioning of remaining cholesterol from the far-HA portion of the envelope into the near-HA nanodomains and partitioning of phospholipids out. This will reduce the size of the nanodomains and permit closer HA-HA approach. The 10 Å change in HA spacing with >90% cholesterol extraction makes this latter nanodomain model more intuitive than the opposite extreme, although either is possible, and indeed the region geometry may change with cholesterol extraction.
Figure 5.
Models for hemagglutinin-lipid interactions in the viral envelope. (a) Regional model of hemagglutinin-lipid interaction, where hemagglutinin partitions into specific regions of the envelope that have a high cholesterol/phospholipid ratio and is excluded from other regions. (White) Cholesterol-enriched regions; (gray) cholesterol-depleted regions. (b) Local model of hemagglutinin-lipid interactions where the hemagglutinin is associated with local lipid domains that have a high cholesterol/phospholipid ratio. The diameter of local domains is <∼10 nm, the most common spacing between hemagglutinin trimers. In either case, depletion of cholesterol (arrows) causes phospholipids to exchange out of the hemagglutinin-containing nanodomains to maintain the cholesterol/phospholipid ratio in those domains. (Triangles) Hemagglutinin trimers. This model does not specify the distribution of neuraminidase tetramers.
Whatever the microscopic basis for the closer spacing of hemagglutinin trimers induced by cholesterol extraction, this structural result helps explain the kinetic response of influenza membrane fusion to membrane cholesterol. We propose that in both influenza virions and simple target liposomes, cholesterol stabilizes lipidic fusion intermediates, reducing the free-energy barrier for formation of stalk and/or early fusion pore structures as proposed previously (25). This accounts for the trend toward decreasing rates of fusion as cholesterol decreases. Simultaneously, we hypothesize that multiple HA trimers must be engaged for efficient fusion (Fig. 1 d)—either via a fixed stoichiometric requirement (10, 11) or via a decreased free-energy barrier with more engaged fusion proteins, as has been shown in analogous synaptic vesicle systems (60, 61).
While the data presented here show that cholesterol extraction increases local HA density and that extraction also speeds fusion of influenza virus with liposomes, several important unknowns remain regarding this process. Although we speculate that higher local HA density at the contact point between virus and liposome may increase the rate of forming lipidic fusion intermediates, the data do not provide sufficient resolution to determine which step in the virus-liposome fusion process is sped by cholesterol extraction. In particular, it is possible that higher local HA density increases the rate of fusion peptide insertion into target liposomes, because this rate will be equal to the number of pH-activated HA at a virus-liposome encounter site times the likelihood per unit time of a single fusion peptide inserting. This could act either alone or in combination with changes to rates of lipidic intermediate formation. In addition, the diffusional mobility of HA in the viral envelope remains unknown; some analyses have approximated HA as immobile on the timescale of fusion (10), while others have treated it as having comparable mobility to that determined for HA in the plasma membrane of transfected cells (9). Although one might reasonably hypothesize that close clustering of HA could actually slow fusion due to steric hindrance of HA conformational change or decrease of lateral mobility, we do not observe such an effect on fusion rates in our measurements. Indeed, as long as cholesterol extraction does not reduce the diffusional mobility of HA in the viral envelope, closer HA-HA spacing driven by cholesterol extraction will increase the probability of multiple engaged HA and therefore speed fusion, as has been previously suggested (62). Both elements of the bimodal kinetic response can thus be explained.
Author Contributions
M.K.D., R.A.D, and K.A.D. performed research, analyzed data, and wrote the article; K.E.Z. performed research and analyzed data; M.Y. designed research and contributed analytic tools; and P.M.K. designed research, contributed analytic tools, analyzed data, and wrote the article.
Acknowledgments
The authors thank C. Stroupe, R. Rawle, A. Radhakrishnan, and B. Ganser-Pornillos for many helpful discussions, and J. White and J. Skehel for the gift of reagents. Transmission electron micrographs were recorded at the University of Virginia Molecular Electron Microscopy Core facility.
This work was supported by National Institutes of Health grants No. R01-GM098304 (to P.M.K.) and No. R01-GM066076 to (M.Y.).
Editor: Joseph Falke.
Footnotes
Four figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00951-0.
Supporting Material
References
- 1.Laver W.G., Valentine R.C. Morphology of the isolated hemagglutinin and neuraminidase subunits of influenza virus. Virology. 1969;38:105–119. doi: 10.1016/0042-6822(69)90132-9. [DOI] [PubMed] [Google Scholar]
- 2.Horne R.W., Waterson A.P., Farnham A.E. The structure and composition of the myxoviruses. I. Electron microscope studies of the structure of myxovirus particles by negative staining techniques. Virology. 1960;11:79–98. doi: 10.1016/0042-6822(60)90056-8. [DOI] [PubMed] [Google Scholar]
- 3.Wrigley N.G. Electron microscopy of influenza virus. Br. Med. Bull. 1979;35:35–38. doi: 10.1093/oxfordjournals.bmb.a071539. [DOI] [PubMed] [Google Scholar]
- 4.Wrigley N.G., Laver W.G., Downie J.C. Binding of antibodies to isolated haemagglutinin and neuraminidase molecules of influenza virus observed in the electron microscope. J. Mol. Biol. 1977;109:405–421. doi: 10.1016/s0022-2836(77)80020-x. [DOI] [PubMed] [Google Scholar]
- 5.Maeda T., Kawasaki K., Ohnishi S. Interaction of influenza virus hemagglutinin with target membrane lipids is a key step in virus-induced hemolysis and fusion at pH 5.2. Proc. Natl. Acad. Sci. USA. 1981;78:4133–4137. doi: 10.1073/pnas.78.7.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang R.T., Wahn K., Rott R. Fusion between cell membrane and liposomes containing the glycoproteins of influenza virus. Virology. 1980;104:294–302. doi: 10.1016/0042-6822(80)90334-7. [DOI] [PubMed] [Google Scholar]
- 7.White J., Kartenbeck J., Helenius A. Membrane fusion activity of influenza virus. EMBO J. 1982;1:217–222. doi: 10.1002/j.1460-2075.1982.tb01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mittal A., Bentz J. Comprehensive kinetic analysis of influenza hemagglutinin-mediated membrane fusion: role of sialate binding. Biophys. J. 2001;81:1521–1535. doi: 10.1016/S0006-3495(01)75806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danieli T., Pelletier S.L., White J.M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanovic T., Choi J.L., Harrison S.C. Influenza-virus membrane fusion by cooperative fold-back of stochastically induced hemagglutinin intermediates. eLife. 2013;2:e00333. doi: 10.7554/eLife.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Floyd D.L., Ragains J.R., van Oijen A.M. Single-particle kinetics of influenza virus membrane fusion. Proc. Natl. Acad. Sci. USA. 2008;105:15382–15387. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kemble G.W., Danieli T., White J.M. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 13.Melikyan G.B., White J.M., Cohen F.S. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J. Cell Biol. 1995;131:679–691. doi: 10.1083/jcb.131.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao H., Armstrong R.T., White J.M. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell. 1999;10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armstrong R.T., Kushnir A.S., White J.M. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cross K.J., Langley W.A., Steinhauer D.A. Composition and functions of the influenza fusion peptide. Protein Pept. Lett. 2009;16:766–778. doi: 10.2174/092986609788681715. [DOI] [PubMed] [Google Scholar]
- 17.Martin J., Wharton S.A., Steinhauer D.A. Studies of the binding of the binding properties of influenza hemagglutinin receptor-site mutants. Virology. 1998;241:101–111. doi: 10.1006/viro.1997.8958. [DOI] [PubMed] [Google Scholar]
- 18.Thoennes S., Li Z.N., Steinhauer D.A. Analysis of residues near the fusion peptide in the influenza hemagglutinin structure for roles in triggering membrane fusion. Virology. 2008;370:403–414. doi: 10.1016/j.virol.2007.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinhauer D.A., Wharton S.A., Wiley D.C. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J. Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun X., Whittaker G.R. Role for influenza virus envelope cholesterol in virus entry and infection. J. Virol. 2003;77:12543–12551. doi: 10.1128/JVI.77.23.12543-12551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munger J., Bennett B.D., Rabinowitz J.D. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda M., Leser G.P., Lamb R.A. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razinkov V.I., Cohen F.S. Sterols and sphingolipids strongly affect the growth of fusion pores induced by the hemagglutinin of influenza virus. Biochemistry. 2000;39:13462–13468. doi: 10.1021/bi0012078. [DOI] [PubMed] [Google Scholar]
- 24.Nussbaum O., Rott R., Loyter A. Fusion of influenza virus particles with liposomes: requirement for cholesterol and virus receptors to allow fusion with and lysis of neutral but not of negatively charged liposomes. J. Gen. Virol. 1992;73:2831–2837. doi: 10.1099/0022-1317-73-11-2831. [DOI] [PubMed] [Google Scholar]
- 25.Biswas S., Yin S.R., Zimmerberg J. Cholesterol promotes hemifusion and pore widening in membrane fusion induced by influenza hemagglutinin. J. Gen. Physiol. 2008;131:503–513. doi: 10.1085/jgp.200709932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerl M.J., Sampaio J.L., Simons K. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 2012;196:213–221. doi: 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domanska M.K., Wrona D., Kasson P.M. Multiphasic effects of cholesterol on influenza fusion kinetics reflect multiple mechanistic roles. Biophys. J. 2013;105:1383–1387. doi: 10.1016/j.bpj.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hess S.T., Kumar M., Zimmerberg J. Quantitative electron microscopy and fluorescence spectroscopy of the membrane distribution of influenza hemagglutinin. J. Cell Biol. 2005;169:965–976. doi: 10.1083/jcb.200412058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scolari S., Engel S., Herrmann A. Lateral distribution of the transmembrane domain of influenza virus hemagglutinin revealed by time-resolved fluorescence imaging. J. Biol. Chem. 2009;284:15708–15716. doi: 10.1074/jbc.M900437200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson R.L., Frisz J.F., Kraft M.L. Hemagglutinin clusters in the plasma membrane are not enriched with cholesterol and sphingolipids. Biophys. J. 2015;108:1652–1659. doi: 10.1016/j.bpj.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheiffele P., Rietveld A., Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 32.Leser G.P., Lamb R.A. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 33.Rossman J.S., Lamb R.A. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris A., Cardone G., Steven A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coorssen J.R., Rand R.P. Effects of cholesterol on the structural transitions induced by diacylglycerol in phosphatidylcholine and phosphatidylethanolamine bilayer systems. Biochem. Cell Biol. 1990;68:65–69. doi: 10.1139/o90-008. [DOI] [PubMed] [Google Scholar]
- 36.García R.A., Pantazatos S.P., MacDonald R.C. Cholesterol stabilizes hemifused phospholipid bilayer vesicles. Biochim. Biophys. Acta Biomembr. 2001;1511:264–270. doi: 10.1016/s0005-2736(01)00283-8. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z., Rand R.P. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophys. J. 1997;73:267–276. doi: 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landsberger F.R., Compans R.W., Lenard J. Organization of the lipid phase in viral membranes. Effects of independent variation of the lipid and the protein composition. Biochemistry. 1973;12:4498–4502. doi: 10.1021/bi00746a030. [DOI] [PubMed] [Google Scholar]
- 39.Scheiffele P., Roth M.G., Simons K. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 1997;16:5501–5508. doi: 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christian A.E., Haynes M.P., Rothblat G.H. Use of cyclodextrins for manipulating cellular cholesterol content. J. Lipid Res. 1997;38:2264–2272. [PubMed] [Google Scholar]
- 41.Klein U., Gimpl G., Fahrenholz F. Alteration of the myometrial plasma membrane cholesterol content with β-cyclodextrin modulates the binding affinity of the oxytocin receptor. Biochemistry. 1995;34:13784–13793. doi: 10.1021/bi00042a009. [DOI] [PubMed] [Google Scholar]
- 42.Yeager M., Berriman J.A., Bellamy A.R. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludtke S.J., Baldwin P.R., Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 44.Tang G., Peng L., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Huby R.D., Dearman R.J., Kimber I. Intracellular phosphotyrosine induction by major histocompatibility complex class II requires co-aggregation with membrane rafts. J. Biol. Chem. 1999;274:22591–22596. doi: 10.1074/jbc.274.32.22591. [DOI] [PubMed] [Google Scholar]
- 46.Kilsdonk E.P., Yancey P.G., Rothblat G.H. Cellular cholesterol efflux mediated by cyclodextrins. J. Biol. Chem. 1995;270:17250–17256. doi: 10.1074/jbc.270.29.17250. [DOI] [PubMed] [Google Scholar]
- 47.Wasilewski S., Calder L.J., Rosenthal P.B. Distribution of surface glycoproteins on influenza A virus determined by electron cryotomography. Vaccine. 2012;30:7368–7373. doi: 10.1016/j.vaccine.2012.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutchinson E.C., Charles P.D., Fodor E. Conserved and host-specific features of influenza virion architecture. Nat. Commun. 2014;5:4816. doi: 10.1038/ncomms5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan J., Tristram-Nagle S., Nagle J.F. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2009;80:021931. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hung W.C., Lee M.T., Huang H.W. The condensing effect of cholesterol in lipid bilayers. Biophys. J. 2007;92:3960–3967. doi: 10.1529/biophysj.106.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stockton G.W., Smith I.C. A deuterium nuclear magnetic resonance study of the condensing effect of cholesterol on egg phosphatidylcholine bilayer membranes. I. Perdeuterated fatty acid probes. Chem. Phys. Lipids. 1976;17:251–263. doi: 10.1016/0009-3084(76)90070-0. [DOI] [PubMed] [Google Scholar]
- 52.Almeida P.F. Thermodynamics of lipid interactions in complex bilayers. Biochim. Biophys. Acta. 2009;1788:72–85. doi: 10.1016/j.bbamem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Heberle F.A., Feigenson G.W. Phase separation in lipid membranes. Cold Spring Harb. Perspect. Biol. 2011 doi: 10.1101/cshperspect.a004630. Published April 1, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McConnell H.M., Radhakrishnan A. Condensed complexes of cholesterol and phospholipids. Biochim. Biophys. Acta. 2003;1610:159–173. doi: 10.1016/s0005-2736(03)00015-4. [DOI] [PubMed] [Google Scholar]
- 55.Radhakrishnan A., McConnell H.M. Condensed complexes of cholesterol and phospholipids. Biophys. J. 1999;77:1507–1517. doi: 10.1016/S0006-3495(99)76998-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itano M.S., Steinhauer C., Jacobson K. Super-resolution imaging of C-type lectin and influenza hemagglutinin nanodomains on plasma membranes using blink microscopy. Biophys. J. 2012;102:1534–1542. doi: 10.1016/j.bpj.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tiffany J.M., Blough H.A. Models of structure of the envelope of influenza virus. Proc. Natl. Acad. Sci. USA. 1970;65:1105–1112. doi: 10.1073/pnas.65.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warren G.B., Houslay M.D., Birdsall N.J. Cholesterol is excluded from the phospholipid annulus surrounding an active calcium transport protein. Nature. 1975;255:684–687. doi: 10.1038/255684a0. [DOI] [PubMed] [Google Scholar]
- 59.Anderson R.G., Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–1825. doi: 10.1126/science.1068886. [DOI] [PubMed] [Google Scholar]
- 60.Domanska M.K., Kiessling V., Tamm L.K. Docking and fast fusion of synaptobrevin vesicles depends on the lipid compositions of the vesicle and the acceptor SNARE complex-containing target membrane. Biophys. J. 2010;99:2936–2946. doi: 10.1016/j.bpj.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hernandez J.M., Kreutzberger A.J., Jahn R. Variable cooperativity in SNARE-mediated membrane fusion. Proc. Natl. Acad. Sci. USA. 2014;111:12037–12042. doi: 10.1073/pnas.1407435111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melikyan G.B., Niles W.D., Cohen F.S. The fusion kinetics of influenza hemagglutinin expressing cells to planar bilayer membranes is affected by HA density and host cell surface. J. Gen. Physiol. 1995;106:783–802. doi: 10.1085/jgp.106.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.