Figure 4.
WEE1 Inhibition Promotes RRM2 Degradation and Aberrant Origin Firing
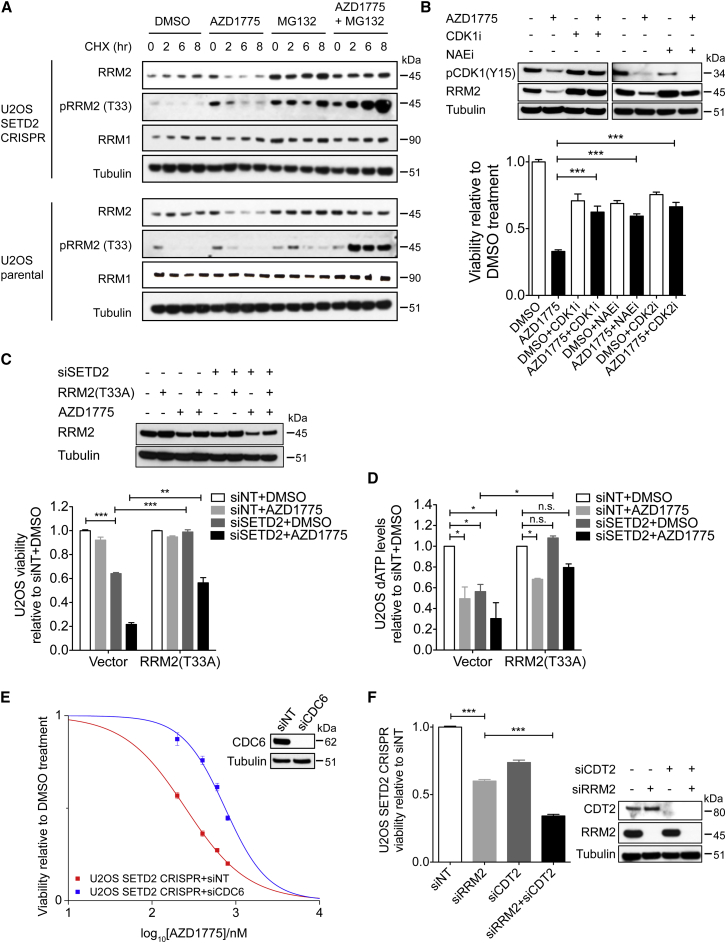
(A) U2OS and U2OS CRISPR SETD2-knockout cells were treated with cycloheximide (50 μg/ml) in the presence of DMSO, AZD1775 (400 nM), MG132 (10 μM) or AZD1775 + MG132. Cells were collected at the indicated times, lysed, and immunoblotted as indicated.
(B) A498 cells were treated with AZD1775 (200 nM) combined with either DMSO, neddylation inhibitor MLN4924 (NAEi) (0.1 mM), CDK1 inhibitor RO3306 (CDK1i) (10 μM) or CDK2 inhibitor (CVT-313) (3 μM). Western blot analysis of phospho-CDK1 (Y15) and RRM2 levels after indicated treatment (24 hr) (top). Viability of A498 cells after indicated treatment (72 hr) (bottom).
(C) U2OS cells expressing either an empty vector (Vector) or a degradation-resistant mutant RRM2 (T33A) were transfected with control siRNA (siNT) or SETD2 siRNA (siSETD2) (48 hr) prior to treatment with either DMSO or AZD1775 (200 nM). Western blot analysis of RRM2 levels after indicated treatment (24 hr) (top), and viability of these cells after indicated treatment (96 hr) (bottom).
(D) dATP levels in U2OS cells stably expressing either an empty vector (Vector) or a degradation-resistant mutant RRM2 (T33A), transfected with control siRNA (siNT) or SETD2 siRNA (siSETD2) (48 hr) prior to treatment with either DMSO or AZD1775 (200 nM) (24 hr). Data are normalized to the control (siNT + DMSO) of each cell line.
(E) Viability curves of U2OS CRISPR SETD2-knockout cells transfected with either control siRNA (siNT) or CDC6 siRNA (siCDC6) (48 hr) and exposed to AZD1775 (5 days) (left). Western blot analysis of CDC6 protein levels (right).
(F) Viability of U2OS CRISPR SETD2-knockout cells transfected with control siRNA (siNT), RRM2 siRNA (siRRM2), CDT2 siRNA (siCDT2), or both (siRRM2 + siCDT2) (48 hr) (left). Western blot analysis of RRM2 and CDT2 protein levels (right). For all graphs in Figure 4, data are presented as mean ± SEM, n = 3 independent experiments. ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; n.s., not significant. Unpaired and two-tailed t tests were used for (B), (C), and (F), and column statistics (one sample t test) were used for (D).
See also Figure S4.