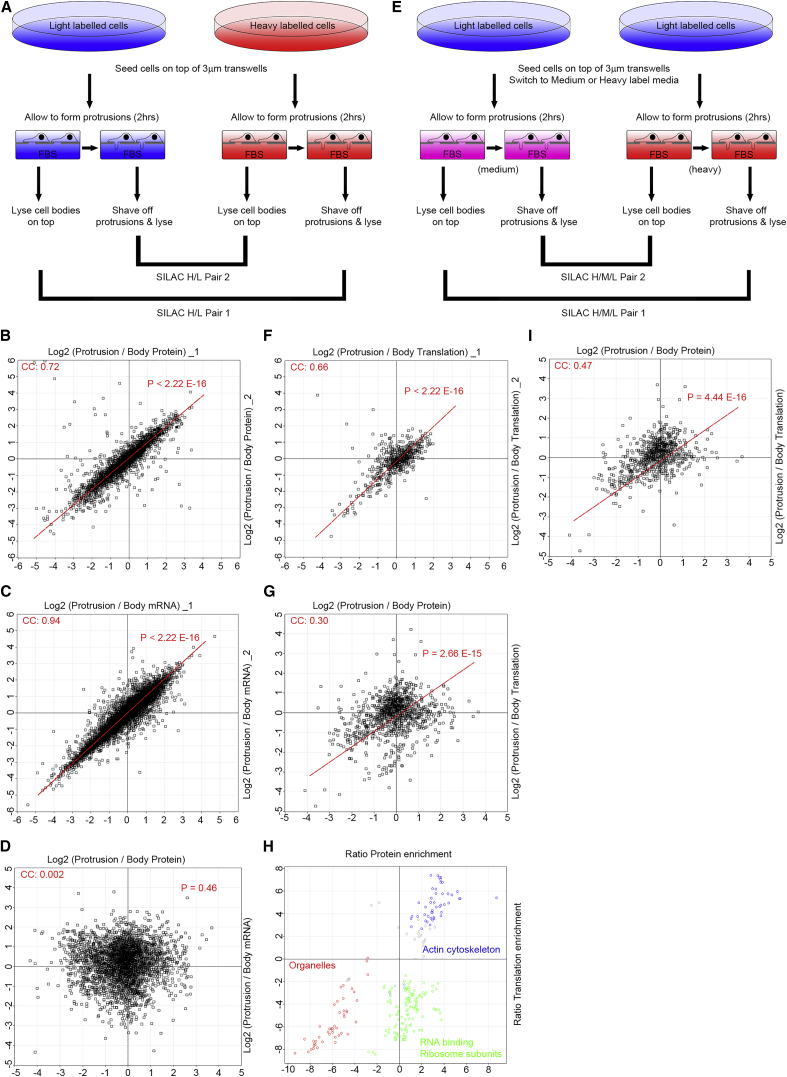
Figure 2.
Localized Translation, but Not mRNA Targeting, Significantly Contributes toward the Proteome Asymmetry between Protrusions and the Cell Body
(A) Schematic representation of SILAC protrusion proteomics analysis; the H/L ratios are the measures of protrusion/body protein distributions.
(B) Proteome distribution between protrusions and the cell body. Log2 SILAC ratios from reciprocal SILAC mixtures of protrusion and cell body fractions (Data S1A) were plotted against each other. CC, Pearson’s correlation coefficient between the two pairs. P value of the correlation is displayed on the graph.
(C) Messenger-RNA distributions between protrusions and the cell body. Log2 of protrusion/cell body FPKM ratios from two replicate experiments (Data S1C) were plotted against each other. CC, Pearson’s correlation coefficient between the two replicates. P value of the correlation is displayed on the graph.
(D) Protein distributions between protrusions and the cell body do not correlate with mRNA distributions. Averaged Log2 of protrusion/cell body protein ratios were plotted against averaged Log2 of Protrusion/Cell body mRNA ratios. CC = Pearson’s correlation coefficient. P-value of the correlation is displayed on the graph.
(E) Schematic representation of pSILAC protrusion proteomics analysis. H/M ratios for each protein are the measures of protrusion/body translation rates.
(F) Translation rate distributions between protrusions and the cell body. Log2 of H/M SILAC ratios from reciprocal pSILAC mixtures of protrusion and cell body fractions (Data S1D) were plotted against each other. CC, Pearson’s correlation coefficient between the two pairs. P value of the correlation is displayed on the graph.
(G) Protein distributions between protrusions and the cell body significantly correlate with translation rate distributions. Averaged Log2 of protrusion/cell body protein ratios from the two reciprocal SILAC pairs were plotted against averaged Log2 of protrusion/cell body pSILAC ratios. CC, Pearson’s correlation coefficient. P value of the correlation is displayed on the graph.
(H) RNA-binding proteins and ribosomal components are enriched but not locally synthesized in protrusions. The 2D-annotation enrichment analysis data from Data S1E was plotted with each data point representing a protein category. Most protein categories exhibit a correlative regulation of their protein distributions and relative translation rates, for example all protrusion-enriched actin-related categories (blue), and all protrusion-depleted organelle-related categories (red), but an anti-correlative behavior is observed for RNA-binding and ribosomal protein categories (green), with their relative translation rates significantly lower than their relative protein amounts.
(I) Removal of RNA-binding and ribosomal protein categories from (G) significantly increases the correlation between protein distributions and translation rates from 0.30 to 0.47. CC, Pearson’s correlation coefficient. P value of the correlation is displayed on the graph.
See also Figure S1.