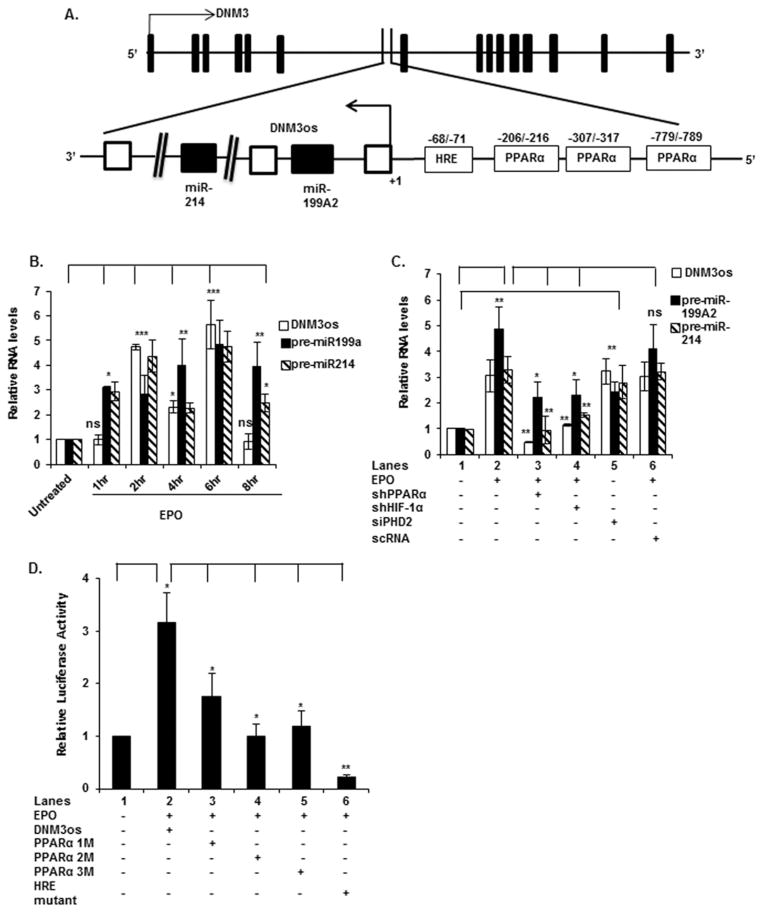
Figure 4. EPO-induced expression of DNM3os RNA, premiR-214 and premiR-199a2 requires the activity of HIF-1α and PPARα.
(A) Schematic diagram of the DNM3os promoter, depicting three binding sites for PPARα and one site for HIF-1α (HRE). Base positions are indicated relative to transcription start site. (B) K562 cells were treated with EPO for the indicated time periods. (C) Cells were transfected with shRNAs for PPARα and HIF-1α followed by EPO treatment for 2 h or transfected with shRNA for PHD2. For (B and C) RNA was subjected to quantitative qRT-PCR utilizing primers listed in Table 1. DNM3os, premiR-199a2 and premiR-214 RNA expression was normalized to GAPDH mRNA levels. qRT-PCR data represent relative levels of DNM3os RNA and premiR-199a2/-214 expression following treatment with EPO compared with untreated cells. (D) Cells were transfected with either the wt DNM3os promoter luciferase construct or the DNM3os promoter constructs with mutations in the PPARα-binding sites at positions − 216/− 206 (PPAR1M), − 317/− 307 (PPAR2M) and − 789/− 779 (PPAR3M) or the HRE site at positions − 71/− 68 (HRE Mut) and treated with EPO for 4 h. Data are expressed as means ± S.D. of three independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05, ns, P>0.05.