Abstract
The intracardiac synthesis of anthracycline alcohol metabolites by aldo-keto reductases (AKRs) contributes to the pathogenesis of anthracycline-related cardiotoxicity. AKR7A2 is the most abundant anthracycline reductase in hearts from donors with and without Down syndrome (DS), and its expression varies between individuals (≈10 fold). We investigated whether DNA methylation impacts AKR7A2 expression in hearts from donors with (n = 11) and without DS (n = 30). Linear models were used to test for associations between methylation status and cardiac AKR7A2 expression. In hearts from donors without DS, DNA methylation status at CpG site -865 correlated with AKR7A2 mRNA (Pearson’s regression coefficient, r = −0.4051, P = 0.0264) and AKR7A2 protein expression (r = −0.5818, P = 0.0071). In heart tissue from donors with DS, DNA methylation status at CpG site −232 correlated with AKR7A2 protein expression (r = 0.8659, P = 0.0025). Multiple linear regression modeling revealed that methylation at several CpG sites is associated with the synthesis of cardiotoxic daunorubicinol. AKR7A2 methylation status in lymphoblastoid cell lines from donors with and without DS was examined to explore potential parallelisms between cardiac tissue and lymphoid cells. These results suggest that DNA methylation impacts AKR7A2 expression and the synthesis of cardiotoxic daunorubicinol.
Keywords: AKR7A2, Cardiotoxicity, DNA Methylation, Heart, Anthracyclines, Down syndrome
Introduction
The use of anthracyclines (e.g., daunorubicin) to treat a variety of cancers is hampered by the development of cardiotoxicity in some patients. For example, a meta-analysis of randomized controlled trials for breast cancer, ovarian cancer, lymphomas and osteosarcomas showed that anthracyclines increased the risk of clinical cardiotoxicity by 5.43-fold, subclinical cardiotoxicity by 6.25-fold, and the risk of cardiac death by 4.94-fold compared with non-anthracycline regimens (1). Risk factors for anthracycline-related cardiotoxicity include total cumulative exposure, younger age at cancer diagnosis, radiation therapy to the chest region, and female sex. Clinical evidence also suggests that Down syndrome status (DS, trisomy 21) increases the risk for cardiotoxicity among patients who receive anthracyclines for the treatment of leukemias (2, 3).
The intracardiac synthesis of C-13 anthracycline alcohol metabolites (e.g., daunorubicinol) by carbonyl reductases (CBRs) and aldo-keto reductases (AKRs) contributes to the complex pathogenesis of anthracycline-related cardiotoxicity (4, 5). The alcohol metabolites synthesized by CBRs and AKRs trigger cardiotoxicity by various mechanisms including inhibition of Ca+2/Mg+2-ATPase in the sarcoplasmic reticulum and inactivation of the cytoplasmic aconitase/iron regulatory protein-1 complex (6, 7).
Recently, we documented the expression of CBRs and AKRs in heart tissue samples from donors with and without DS (8). Our findings suggest that cardiac CBR1, AKR1A1, and AKR7A2 protein levels are significant contributors to cardiac daunorubicin reductase activity. Of note, AKR7A2 appears to be the most abundant anthracycline reductase accounting for approximately 36% of the total reductase content in hearts from donors with and without DS. Cardiac AKR7A2 expression showed considerable variability at the mRNA (DS: 13-fold, and non-DS: 89-fold) and protein levels (DS: 9-fold, and non-DS: 13-fold).
Developments in the field of epigenomics are demonstrating that DNA methylation can affect the pharmacodynamics of various drugs by modulating the expression of specific drug metabolizing enzymes (9). In this context, it is reasonable to hypothesize that interindividual variability in cardiac AKR7A2 gene expression may impact the metabolism of anthracycline substrates and consequently the risk of cardiotoxicity. Thus, the main goal of this study was to perform quantitative DNA methylation analysis on selected regions of the AKR7A2 locus by using cardiac DNA isolated from donors with and without DS. We also investigated the impact of DNA methylation status at different sites on cardiac AKR7A2 gene expression (mRNA and protein) and on cardiac daunorubicin reductase activity. It’s is not known whether accessible “proxy” tissues and/or cells (e.g., peripheral blood lymphocytes) are useful surrogates for predicting the intracardiac synthesis of cardiotoxic anthracycline alcohol metabolites. This knowledge may contribute to propel further research with potential practical implications for predicting the risk of cardiotoxicity. For example, one can envision a diagnostic scenario in which the maximal synthesis rates of doxorubicinol in the heart tissue of cancer patients undergoing chemotherapy with anthracyclines are inferred from a readily accessible “proxy” such as peripheral blood lymphocytes trough inter-tissue extrapolation approaches (10, 11). Peripheral blood lymphocytes have substantial CBR/AKR activity toward anthracycline substrates (12, 13). This is because lymphocytes express CBRs and AKRs, including AKR7A2 (14, 15). Thus, pilot studies in lymphoblastoid cell lines derived from donors with and without DS were performed to examine the dynamics of methylation at the AKR7A2 locus.
Materials and Methods
Heart samples
The Institutional Review Board of the State University of New York at Buffalo approved this research. Heart samples from donors with (n = 11) and without DS (n = 30) were procured from The National Disease Research Interchange (NDRI, funded by the National Center for Research Resources), The Cooperative Human Tissue Network (CHTN, funded by the National Cancer Institute), and The National Institute of Child Health and Human Development (NICHD) Brain and Tissue Bank. The postmortem to tissue recovery interval was ≤10 h. Samples (2 – 20 g, myocardium, left ventricle only) were frozen immediately after recovery and stored in liquid nitrogen until further processing. The main demographics of heart donors are summarized in Table 1 (16). Down syndrome status (yes/no) was obtained from anonymous medical histories and confirmed by array comparative genomic hybridization (aCGH) as described (4, 8, 16). Cardiac DNA, RNA and cytosolic fractions were isolated by following standardized procedures as described (4, 8, 17).
Table 1.
Demographics of heart donors
| Non-DS | DS | |
|---|---|---|
| Number of subjects | 30 | 11 |
| Mean age (years) | 63.1 | 39.0 |
| Standard deviation of age | 20.2 | 28.8 |
| Males | 13 | 10 |
| Females | 17 | 1 |
| Black Subjects | 7 | 6 |
| White Subjects | 2 | 24 |
Cell culture and treatments
B-lymphoblastoid cell lines derived from donors with (GM01921 and AG09394) and without DS (GM18856 and GM18870) were obtained from the Coriell Cell Repository (Camden, NJ). Cell cultures were grown in RPMI 1640 media (Invitrogen, Carlsbad, CA) containing heat inactivated fetal bovine serum (15% v/v, Invitrogen), penicillin (100 U/ml, Invitrogen) and streptomycin (100 U/ml, Invitrogen). Cells were grown in standard incubator conditions at 37°C, 5% CO2, and 95% relative humidity. During experiments, cells were initially seeded into T-25 flasks at a density of 2.75 × 105 cells/ml. On Day 1, cells were treated with the demethylating drug 5’Aza-2’Deoxycytidine (1 µM. EMD Millipore, Darmstadt, Germany) or dimethyl sulfoxide vehicle (< 1%. ATCC, Manassas, VA). Cells were harvested at 12 and 72 h after treatments for isolation of DNA and RNA with E.Z.N.A. DNA/RNA Isolation Kits (Omega Bio-TK, Norcross, GA).
Quantitative DNA methylation analysis
Targeted methylation analysis was performed with a Sequenom MassARRAY EpiTyper (Sequenom, San Diego, CA) at the Genomics Shared Resource, Roswell Park Cancer Institute (Buffalo, NY). Briefly, genomic DNA (gDNA) from cardiac tissue was bisulfite converted using an EZ Bisulfite conversion kit according to manufacturer’s instructions (Zymo Research, Irvine CA). Bisulfite converted primers were designed to cover the region of interest in AKR7A2 using Sequenom’s primer design website (http://www.epidesigner.com). The sets of primers covered approximately 1.6 kb upstream the translation initiation codon of AKR7A2 (supplemental Table 1, Figure 1). During PCR amplification a T7 promoter-tagged reverse primer was incorporated into the amplification product to act as an anti-sense primer tag for in vitro transcription, and preserve the bisulfite changes. Amplification products were treated with shrimp alkaline phosphatase and T-Cleavage transcription/RNase A cocktail.
Figure 1.
Top: quantitative DNA methylation analysis of the AKR7A2 locus in human heart samples from donors with (black boxes) and without DS (grey boxes). The positions of methylated sites are relative to the translation initiation codon (A+1TG). Boxes show the 25th and 75th percentile, and whiskers indicate the 5th and 95th percentile. Horizontal lines indicate means and dots represent outliers. Bottom: schematic representation of the 5’ region of AKR7A2 annotated with potential consensus motifs for transcription factors.
Cleavage products, and appropriate genomic DNA standards (0%, 25%, 50%, 100% methylated) were analyzed by Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry. Non-methylated and methylated cytosines generated different mass signals, and the intensities were compared to determine methylated/non-methylated ratios. Relative methylation analysis for each CpG site was performed with Sequenom’s proprietary software in order to produce detailed quantitative “epigraphs” and MS Excel files (Microsoft, Redmond, WA) for subsequent data analysis.
AKR7A2 mRNA and protein expression
Cellular AKR7A2 mRNA expression was analyzed by quantitative real-time PCR with specific primers by following the MIQE guidelines (18). The primers for AKR7A2 (forward: 5’-GGCCTCTCCAACTATGCTAG-3’, and reverse: 5’-GGCATAGAACCTCAGTCCAAAG-3’) were obtained from realtimeprimers.com (Elkins Park, PA). Cardiac AKR7A2 protein expression was determined by quantitative immunoblotting with a specific anti-AKR7A2 antibody (Abnova, Taipei City, Taiwan) and recombinant AKR7A2 protein standards. Cardiac AKR7A2 mRNA and AKR7A2 protein expression in samples from donors with and without DS have been recently reported by us (8).
Cell cycle analysis
Cell cycle analysis was performed by measuring DNA content with propidium iodide in a FACSCalibur 1 flow cytometer (Becton Dickinson, Franklin Lakes, NJ) at the Department of Flow and Image Cytometry (Roswell Park Cancer Institute), essentially as described (19).
Statistical Data Analysis
Analyses were performed using GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA). The Kolmogorov-Smirnov test was used to determine the normality of data sets. The Mann-Whitney U test or Student’s t-test were used to compare group means. ANOVA with Tukey’s post hoc analysis was used to analyze differences in methylation sites. Pearson's and Spearman’s coefficients of correlation were used to analyze data sets with normal and non-normal distributions respectively. Statistical comparisons were considered significant at P < 0.05.
Multiple linear regression models were fit using the R programming language (http://www.r-project.org/). Two different types of models were fit in order to: (1) estimate AKR7A2 mRNA normalized expression as a function of different methylation sites, and (2) determine whether DNA methylation in AKR7A2 is associated with the cardiac synthesis rates of the cardiotoxic anthracycline alcohol metabolite daunorubicinol. The methylation sites were filtered to include only those sites with means that exceeded the lower limit of detection of 0.10 (i.e., > 10% methylation. Filtering left five sites as candidate predictors in the various regression models (Figure 1). The fitted models for AKR7A2 mRNA expression are of the form:
AKR7A2mRNA = β0 + β1 · Sex · + β2 · Site1 + ⋯ + βk+1 · Sitek +ε
where β0 is the intercept term, Sex is a two-level factor and Sitek represents methylation site k, and ε is the error term.
In order to assess the impact of DNA methylation in AKR7A2 on cardiac daunorubicin reductase activity (DA), we leveraged a recent study describing the expression of anthracycline reductases, including AKR7A2, in the same collection of heart samples (8). In this work, a multiple linear regression model was developed in which the original AKR7A2 “protein term” (i.e., absolute AKR7A2 protein abundance expressed as nmol/g cytosolic protein) was replaced by terms including methylation status at the differentially methylated sites in the AKR7A2 locus. The protein levels were transformed to a log10 scale before model fitting. The fitted models are of the general form:
DA = β0 + β1 · Sex + β2 · Condition + β3 · CRB1 + β4 · AKR1A1 + βs · Sitek + ⋯ + βk+5 · Sitek + ε
where DA is maximal daunorubicin reductase activity in heart cytosols, Condition is a two-level factor indicating DS and non-DS states, and the other terms are as previously defined in the AKR7A2 mRNA model. Another multiple regression model of this type was fit to only non-DS samples. Note that the sample size did not support models of only DS samples.
Hypotheses test on the regression coefficients were performed for the different models to determine the significance of each term in predicting the response variables (AKR7A2 mRNA expression and daunorubicin reductase activity). In order to identify the most parsimonious model that explains the different responses, a drop-one approach was applied, which entails model reduction through the elimination of less significant factors (20).
Results
AKR7A2 methylation status in hearts from donors with and without DS
Candidate methylation sites in the region encompassing up to 1.6 kilobases upstream the translation initiation codon of AKR7A2 were identified based on: a) high overall CpG content, and b) overlap with binding motifs for potentially relevant transcription factors. Putative binding motifs for the AKR7A2 promoter region were identified with a combination of algorithms including Alggen, Transfac, and MatInspector (21, 22). The region of interest included CpG islands and CpG “shores” (23). A CpG “shore” is a region that is close to a CpG island, but which contains relatively fewer CpG sites. Only sites with methylation means above the lower limit of detection of 0.1 (i.e., > 10% methylation) were included for analysis. Figure 1 shows the quantitative DNA methylation profile of AKR7A2 in cardiac DNA (left ventricle) isolated from donors with (n = 11) and without DS (n = 30). Highly methylated sites exhibited variable methylation between samples (site −877, range: 0.17 – 0.52, site −865, range: 0.29 – 0.71, site −852, range: 0.0 – 0.47, and site −232, range: 0.15 – 0.64). Sites −877, −865 and −852 form a CpG methylation block that is 120 bp upstream a non-validated xenobiotic response element motif (XRE); further comparisons showed a significant difference between the averages of methylation at sites −877, −865 and −852, respectively (P < 0.0001) (24). Additional statistical comparison tests were performed after the stratification of samples by DS status (Figure 1). The average methylation at site −852 was 24% lower in samples from donors without DS than in samples from donors with DS, although the difference between means fell just below the significance threshold (P = 0.0602). Likewise, there were no significant differences in average methylation status at sites −877 and −865 between samples from donors with and without DS (P > 0.05. Figure 1). The levels of DNA methylation at specific loci may differ between individuals with African and European ancestry (25). There were no significant differences in methylation status at the AKR7A2 sites −877, −865, and −852 between cardiac DNA samples from self-reported black (i.e., African) and white (i.e., European) donors (P > 0.05. Supplemental figure S1). There were no significant associations between donor’s age and AKR7A2 methylation status at sites −877, −865, and −852 (Supplemental figure S2). AKR7A2 methylation status at sites −877 and −865 were similar in female and male donors. AKR7A2 methylation status at site −852 was 7.2% higher in cardiac DNA from males than in DNA from female donors (P < 0.05. Supplemental figure S3).
Cardiac AKR7A2 methylation and gene expression
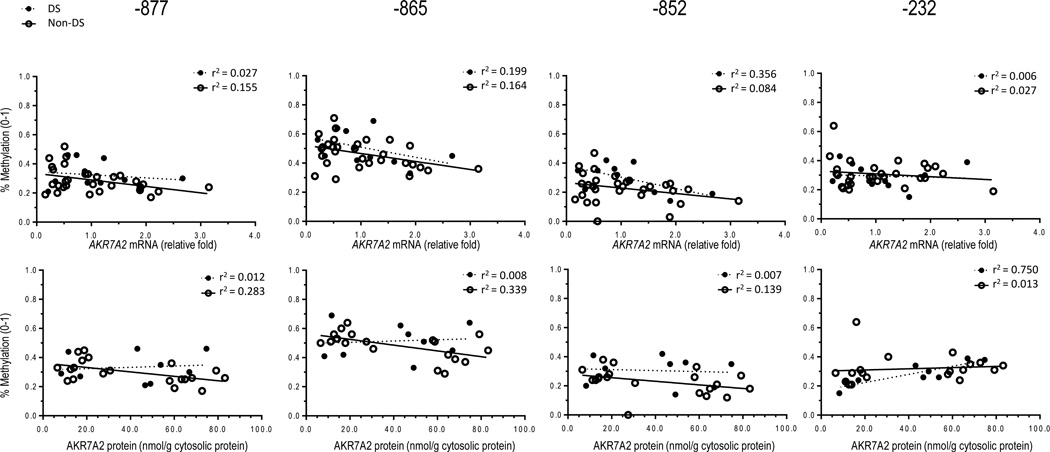
Correlation analyses were performed to determine whether DNA methylation is associated with AKR7A2 gene expression in heart tissue from donors with an without DS. Methylation status at CpG sites −877 and −852 did not show significant correlation with cardiac AKR7A2 expression at the mRNA and protein levels in samples from donors without DS (Figure 2). However, DNA methylation status at CpG site −865 was significantly associated with AKR7A2 mRNA levels (Pearson’s correlation coefficient, r = −0.4051, P = 0.0264) and AKR7A2 protein expression (Pearson’s correlation coefficient, r = −0.5818, P = 0.0071) in hearts from donors without DS. Methylation in CpG site −865 accounts for up to 16% and 34% of the variabilty in cardiac AKR7A2 mRNA and protein expression, respectively (Figure 2). Methylation at sites −877, −865, and −852 was not significantly associated with AKR7A2 expression (mRNA and protein) in heart samples from donors with DS (P > 0.05) (Figure 2). In samples from donors with DS, the extent of methylation at site −232 was significantly associated with cardiac AKR7A2 protein expression (Pearson’s correlation coefficient, r = 0.8659, P = 0.0025). Furthermore, the univariate regression analysis suggests that up to 75% of the variability in cardiac AKR7A2 protein expression can be attributed to the extent of methylation at site −232 (Figure 2). There were no significant associations detected between methylation status at site −232 and cardiac AKR7A2 expression in heart samples from donors without DS (Figure 2).
Figure 2.
Top: linear regression analyses of percent methylation at variable sites in AKR7A2 versus cardiac AKR7A2 mRNA expression. Each circle depicts the average of individual samples analyzed in triplicates. Bottom: linear regression analyses of percent methylation at variable sites in AKR7A2 versus cardiac AKR7A2 protein expression in donors with and without DS. Each circle depicts the average of individual samples analyzed in triplicates.
Independent multiple linear regressions were performed to further determine the contribution of differentially methylated sites to cardiac AKR7A2 gene expression. Results suggest that methylation status at site −852 is a significant factor for cardiac AKR7A2 mRNA expression in samples from donors without DS, whereas methylation status at site −232 is significantly associated with AKR7A2 mRNA expression in samples from donors with DS (Table 2).
Table 2.
The most parsimonious models for the multiple regression of normalized AKR7A2mRNA expression including methylation sites in heart samples from donors without (top) and with DS (bottom). This model was obtained from a full model containing five candidate methylation sites, and reduced using a drop-one approach until only significant factors remained. The columns represent the coefficients, standard error of the coefficient estimates, t-test and p-values from the hypothesis test on the coefficients.
| Non-DS | ||||
| Term | β-estimate | Std. Error | t-stat | P-Value |
| (Intercept) | 4.697 | 1.011 | 4.648 | 0.000315 |
| CpG−865 | −5.813 | 2.019 | −2.879 | 0.011476* |
| DS | ||||
| Term | β-estimate | Std. Error | t-stat | P-Value |
| (Intercept) | −1.0870 | 0.6712 | −1.620 | 0.15645 |
| CpG−232 | 9.3224 | 2.3862 | 3.907 | 0.00792* |
Next, we investigated whether DNA methylation in AKR7A2 is associated with the cardiac synthesis rates of the cardiotoxic anthracycline alcohol metabolite daunorubicinol. We used datasets from our recent study describing the expression of anthracycline reductases, including AKR7A2, in the same collection of heart samples (8). In order to simultaneously estimate the collective impact of the methylated sites in AKR7A2 on daunorubicin reductase activity, a multiple linear regression model was developed in which the original AKR7A2 “protein term” (i.e., absolute AKR7A2 protein abundance expressed as nmol/g cytosolic protein) was replaced by terms including methylation status at the differentially methylated sites. Multiple regression is complementary to the univariate analyses and significant factors in the univariate and multiple regression models may not be in agreement (26). CBR1 and AKR1A1 represent protein abundance in heart (nmol/g cytosolic protein) and were included as predictors in the modeling (8). Hypotheses tests on the regression coefficients revealed that DNA methylation at sites −865 and −852 are significant contributors to maximal daunorubicin reductase activity in heart tissue (Tables 3 and 4).
Table 3.
Results from the multiple regression of cardiac daunorubicinactivity incorporating CRB1 and AKR1A1 protein levels, as well as AKR7A2methylation status for sites −865 and −852 for samples from donors with and without DS. The columns represent the coefficients, standard error of the coefficient estimates, t-test and p-values from the hypothesis test on the coefficients.
| Term | β estimate | Std. Error | t-stat | P-value |
|---|---|---|---|---|
| Intercept | 2.6015 | 1.6760 | 1.552 | 0.139026 |
| Condition | 1.3440 | 0.6603 | 2.036 | 0.057693 |
| CBR1 | 1.9217 | 0.6790 | 2.830 | 0.011551 |
| AKR1A1 | 1.2925 | 0.5739 | 2.252 | 0.037823 |
| −865 | −19.5980 | 4.8270 | −4.060 | 0.000814 |
| −852 | 19.5696 | 5.9876 | 3.268 | 0.004529 |
Table 4.
Results from the multiple regression of cardiac daunorubicinactivity incorporating CRB1 and AKR1A1 protein levels, as well as AKR7A2methylation status for sites −865 and −852 for samples from donors without DS. The columns represent the coefficients, standard error of the coefficient estimates, t-test and p-values from the hypothesis test on the coefficients.
| Term | β estimate | Std. Error | t-stat | P-value |
|---|---|---|---|---|
| Intercept | 4.4153 | 1.6474 | 2.680 | 0.01889 |
| CBR1 | 1.8666 | 0.7232 | 2.581 | 0.02281 |
| AKR1A1 | 1.4872 | 0.6278 | 2.369 | 0.03400 |
| −865 | −18.3181 | 5.8102 | −3.153 | 0.00763 |
| −852 | 14.5524 | 6.9928 | 2.081 | 0.05776 |
AKR7A2 methylation analysis in lymphoblastoid cell lines from donors with and without DS
To analyze the dynamics of AKR7A2 gene methylation in a potential “proxy” cell type, lymphoblastoid cell lines derived from individual donors with and without DS were treated with the demethylating drug 5’Aza-2’Deoxycytidine (DAC) followed by quantitative DNA methylation analysis. The effect of DAC treatment on the cell cycle was also investigated. In the two cell lines derived from donors without DS, DAC treatment for 12 h did not impact the extent of methylation at sites −877, −865, −852 and −232, in comparison to cultures incubated with DMSO vehicle (Figure 3). The cell cycle profiles at 12 hr of DAC treated cells without DS were similar to control cultures (Table 5). In the cells without DS, longer treatment with DAC (72 h) decreased the extent of methylation in the CpG block containing sites −877, −865, and −852, and did not modify methylation status at site −232 (Figure 3 and supplemental figure S4). The 72 h DAC treatment induced cell cycle arrest at the G0/G1 phase and decreased the fraction of cells in the S phase (Table 5 and Supplemental figure S5).
Figure 3.
Quantitative DNA methylation analyses of the AKR7A2 locus in lymphoblastoid cell lines derived from donors with (GM01921) and without DS (GM18856). Top: cell lines were treated with DAC (black circles) or DMSO vehicle (white circles) for 12 h. Bottom: cell lines were treated with DAC (black circles) or DMSO vehicle (white circles) for 72 h. Data represent the mean ± SD of two individual experiments performed in duplicates. * P >0.05 at differentially methylated sites. ** P < 0.05 at differentially methylated sites.
Table 5.
Cell cycle analysis in lymphoblastoid cell lines treated with vehicle (DMSO) and DAC. Each value represent the mean from two experiments performed in parallel.
| Non-Down syndrome | Down syndrome | |||||||
|---|---|---|---|---|---|---|---|---|
| 12 hr DMSO |
72 hr DMSO |
12 hr DAC |
72 hr DAC |
12 hr DMSO |
72 hr DMSO |
12 hr DAC |
72 hr DAC |
|
| %G0/G1 | 64.4 | 75.2 | 66.1 | 81.8 | 65.2 | 68.7 | 66.6 | 82.6 |
| %G2/M | 8.4 | 7.5 | 8.7 | 6.2 | 6.0 | 8.5 | 5.9 | 6.3 |
| %S | 27.1 | 17.3 | 25.2 | 12.0 | 28.9 | 22.8 | 27.5 | 11.2 |
| CV | 5.5 | 5.4 | 5.3 | 6.0 | 7 | 6.4 | 7.3 | 7.2 |
Cell lines derived from donors with DS did not exhibit significant changes in methylation status at sites −877, −852 and −232 after the 12 h treatment with DAC (P > 0.05) (Figure 3 and supplemental figure S4). The 12 h treatment did not induce changes in the cell cycle profile in comparison to control cultures (Table 5). The 72 h treatment with DAC tended to decrease the extent of methylation in sites −877, −865, and −852 in both lines from donors with DS, but only cell line AG09394 showed a significant decrease in methylation at site −865. The extent of methylation at site −232 decreased significantly in cell line GM01921, but showed no significant changes in cell line AG09394 (Figure 3 and supplemental figure S4). In cells with DS, the 72 h DAC treatment induced in cell cycle arrest at the G0/G1 phase in a manner similar to cells without DS (Table 5 and Supplemental figure S5).
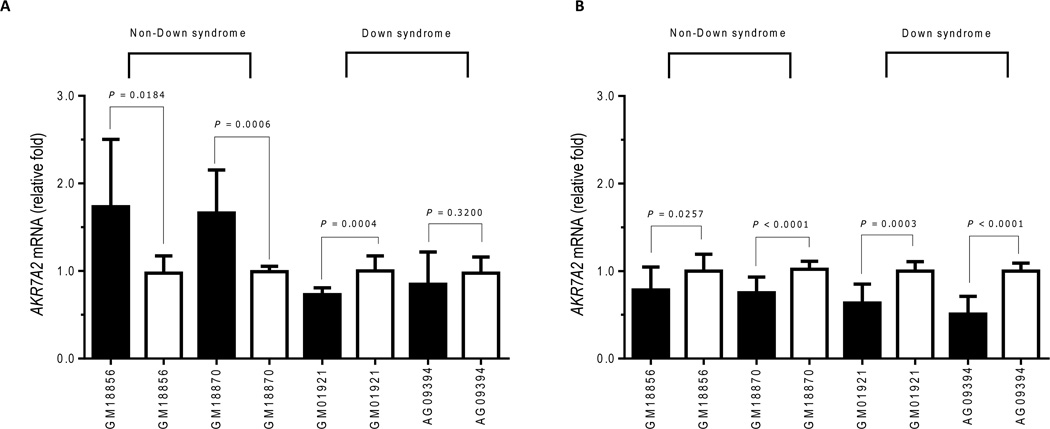
Treatment with DAC for 12 hours significantly increased the expression of AKR7A2 mRNA in the two cell lines derived from donors without DS (Figure 4). In both cell lines, the 72 h treatment with DAC decreased the expression of AKR7A2 mRNA in comparison to incubations with the DMSO vehicle (Figure 4). The two lines from donors with DS exhibited different AKR7A2 mRNA expression in response to treatments with DAC. That is, AKR7A2 mRNA expression decreased in cell line GM01921 after the 12 h treatment with DAC, whereas AKR7A2 mRNA expression in cell line AG09394 did not change after the 12 h treatment with DAC (Figure 4). Treatment with DAC for 72 h decreased the expression of AKR7A2 mRNA in both lines from donors with DS (Figure 4).
Figure 4.
Effect of DAC on AKR7A2 mRNA expression in lymphoblastoid cell lines derived from donors with and without DS. Cell lines were treated with DAC (black bars) or DMSO vehicle (clear bars) for 12 h (panel A) or 72 h (panel B). Data represent the mean ± SD of two individual experiments performed in duplicates.
Discussion
Variability in gene expression is due to the complex interplay between genetic and epigenetic factors that are in turn affected by endogenous and exogenous influences. There is a relative paucity of reports assessing the impact of DNA methylation on the expression of genes encoding for drug metabolizing enzymes in human heart. Thus, the main goal of this study was to investigate the impact of DNA methylation on the variable cardiac expression of AKR7A2, the most abundant anthracycline reductase in hearts from donors with and without DS. The relatively high linear correlations between DNA methylation status in specific CpG sites of the AKR7A2 locus (i.e., sites −877, −865, −852, and −232) suggest potentially relevant associations with: a) cardiac AKR7A2 gene expression, and b) maximal daunorubicin reductase activity. The magnitudes of the observed linear correlations represent the fraction of variability in total cardiac AKR7A2 expression explained by DNA methylation at each specific site. These values are well in line with recent reports describing the effect of DNA methylation status on the expression of various functional loci including the one encoding the prominent drug metabolizing enzyme CYP3A4 (24, 27, 28). Thus, it appears that DNA methylation is a genuine co-variable accounting for a fraction of the variability observed for cardiac AKR7A2 expression (Figures 1 and 2).
Our results also suggest that the impact of methylated sites on cardiac AKR7A2 gene expression is different in samples from donors with and without DS. That is, methylation status at site −865 is variable and impacts cardiac mRNA and protein expression in heart tissue from donors without DS (Figures 1 and 2). These observations are further supported by the significant associations identified for site −865 through independent multiple linear regression models. Specifically, the models suggest that quantitative methylation status at site −865 is significantly associated with AKR7A2 mRNA expression and maximal daunorubicin reductase activity in heart tissue from donors without DS (Tables 2 and 3). The relative proximity between the CpG block constituted by sites −877, −865 and −852 and a non-validated XRE motif in the proximal promoter region is in line with evidence describing the presence of regulatory CpGs near the binding sites for methylation sensitive transcription factors such as the aryl hydrocarbon receptor (AHR) (29). In this regard, it would be interesting to test whether AKR7A2 gene expression is regulated by AHR through the conserved XRE motif in the context of variable methylation status at sites −877, −865 and −852.
The differentially methylated site −232 appears to impact AKR7A2 gene expression in heart samples from donors with DS. Although these observations are limited by the small sample size, there was a relatively strong positive association (r2 = 0.75) between methylation status at site −232 and cardiac AKR7A2 protein expression in heart tissue from donors with DS. In agreement, the exploratory multiple regression modeling restricted to mRNA data suggests that the extent of methylation in site −232 is positively associated with cardiac AKR7A2 mRNA expression in the DS setting. The positive sign of the association is intriguing because, in general, DNA methylation causes transcriptional silencing. Recently, Letourneau et al. reported a global increase in chromatin accessibility in DS, which tends to flatten gene expression levels at the genome-wide scale (30, 31). Thus, it’s possible to speculate that methylation at site −232 may constitute a proxy for a state of increased chromatin accessibility in the gene promoter region of AKR7A2, which would in turn favor gene transcription by diluting the repressing effect of DNA methylation.
Adkins et al. analyzed approximately 27,000 autosomal CpG sites assigned to 14,500 loci and found that 13.7% of the CpGs exhibited different amounts of DNA methylation between subjects of self-reported African and European ancestries (25). To the best of our knowledge, the extent of methylation in the AKR7A2 locus in subjects with diverse geographical ancestries has not been described. Although our comparison is somewhat limited by the small sample size, analysis of the 95% confidence intervals at each CpG site suggests that the extent of DNA methylation at sites −877, −865 and −852 in AKR7A2 is similar in cardiac tissue from black and white donors (32) (Supplemental figure S1). The extent of methylation at AKR7A2 site −852 was higher in males than in females (Supplemental figure S3), and multiple linear regression suggests that methylation status at site −852 impacts the synthesis of cardiotoxic daunorubicinol (Table 4). However, the potential impact of differential methylation by gender at each CpG site on cardiac daunorubicin reductase activity could not be investigated through multiple linear regression procedures due to sample size limitations. This avenue deserves further consideration given the documented differences in terms of risk of anthracycline-related cardiotoxicity between male and female cancer survivors (33–35).
We also attempted to define whether DNA methylation impacts AKR7A2 expression in lymphoblastoid cell lines derived from donors with and without DS. Our aim was to set the foundation for future studies focused on exploring whether information derived from “proxy” cells (e.g., lymphocytes from peripheral blood) would be useful to predict the cardiac synthesis of toxic anthracycline metabolites (i.e., metabolic clearance) through inter-tissue extrapolation approaches. The expression of AKR7A2 mRNA was induced in lymphoblastoid cells from donors without DS after 12 h treatment with the de-methylating agent DAC; this phenomenon is consistent with gene regulation through DNA methylation. However, prolonged treatment with DAC did not significantly increase cellular AKR7A2 mRNA levels. In general, the extent of DNA methylation at sites −877, −865, and −852 changed after the 72 h exposure to DAC, although the magnitude of the changes varied between individual cell lines (Figure 3). Lymphoblastoid cells have a doubling time of approximately 36 h, and previous studies have shown that relatively prolonged treatments with DAC (e.g., 1 – 5 µM, 72 h) induce cell cycle arrest at the G0/G1 phase (36, 37). In agreement, our results show that: a) lymphoblastoid cells from donors with and without DS exhibit cell cycle profiles similar to control incubations after the 12 h treatment with DAC, and b) the 72 treatment with DAC increases the proportion of cells at the G0/G1 phase in DS and non-DS cells (Table 5 and supplemental figure S3). The observed decreases in DNA methylation in AKR7A2 after the 72 h time point is consistent with the synthesis of non-methylated DNA due to the covalent binding of DAC to DNA methyltransferase. In this regard, the cell cycle analysis show that approximately 11% of the cells are in the S phase. The observed decreases in AKR7A2 mRNA levels in DS and non-DS cells may be due to the transcriptional repression of AKR7A2 in the fraction of cells arrested at the G0/G1 phase of the cell cycle (~80%. Table 5 and supplemental figure 3). Additional experiments would be needed to determine whether the dynamics of AKR7A2 gene expression are impacted by quantitative changes in the cell cycle. However, the fact that specific sites in the AKR7A2 locus (e.g., −877, −865, and −852) were differentially methylated in tissue samples made of terminally differentiated cells (i.e., human myocardium) as well as in samples from rapidly dividing cells (i.e., lymphoblastoid cells), points to the potential relevance of the CpG sites for regulating AKR7A2 gene expression in different cellular contexts. Thus, it will be interesting to examine: a) whether the extent of methylation at the prominent sites in AKR7A2 correlate between heart tissue and peripheral blood lymphocytes isolated from the same donor (i.e., paired), and b) whether DNA methylation status impacts AKR7A2 expression and consequently daunorubicin reductase activity in lymphocytes. If successful, these efforts would be critical to define the appropriate scalars for the non-cardiac-to-cardiac tissue extrapolation of maximal daunorubicinol synthesis rates (10, 11).
This study is limited by the small number of cardiac samples from donors with DS. The scarcity of tissue samples from donors with DS hampers research on fundamental topics related to the metabolism and disposition of drugs in the DS setting. Our procurement rates for samples from donors with DS is low (approximately 1 sample every 9 - 12 months), even after working with national cooperative resources such as NDRI, CHTN, and the NICHD Brain and Tissue Bank. Also, determinations of the effect size of potential co-variables (e.g., medication use, concomitant diseases) on cardiac AKR7A2 methylation status were hindered by the combined impact of sample size limitations and incomplete clinical records from some donors. In general, the level of detail included in the heart samples’ clinical background information is insufficient to conduct additional analyses. Nevertheless, the group of heart samples analyzed in this study may contribute informative data to define tissue-specific determinants for anthracycline-related cardiotoxicity. The complex pathogenesis of anthracycline-related cardiotoxicity is impacted by modifiable (e.g., obesity, metabolic syndrome) and non-modifiable risk factors (e.g., Down syndrome status, younger age) (38). Recent evidence, suggests that the list of non-modifiable risk factors could be expanded by including specific single nucleotide polymorphic variants that have been associated with the risk of anthracycline-related cardiotoxicity through genetic association studies (39–42). Findings from the present study suggest that the intracardiac synthesis of cardiotoxic alcohol metabolites may be also impacted by DNA methylation status at specific sites in the AKR7A2 locus. We therefore propose that consideration be given to the potential role of epigenetic modifications in the network of genes associated with the variable pharmacodynamics of anthracyclines. Further understanding of such epigenetic modifications could contribute to the discovery of novel determinants for the risk of cardiotoxicity.
Supplementary Material
Acknowledgments
Research in this report was supported by the National Institute of General Medical Sciences and the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under awards R01GM073646 and R03HD076055. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Carrie C. Hoefer, Department of Pharmaceutical Sciences, The State University of New York at Buffalo, Buffalo, NY 14260, USA
Adolfo Quiñones-Lombraña, Department of Pharmaceutical Sciences, The State University of New York at Buffalo, Buffalo, NY 14260, USA.
Rachael Hageman Blair, Department of Biostatistics, School of Public Health and Health Professions, The State University of New York at Buffalo, Buffalo, New York, 14260, USA.
Javier G. Blanco, Department of Pharmaceutical Sciences, The State University of New York at Buffalo, Buffalo, NY 14260, USA
References
- 1.Smith LA, Cornelius VR, Plummer CJ, Levitt G, Verrill M, Canney P, Jones A. Cardiotoxicity of anthracycline agents for the treatment of cancer: systematic review and meta-analysis of randomised controlled trials. BMC Cancer. 2010;10:337. doi: 10.1186/1471-2407-10-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krischer JP, Epstein S, Cuthbertson DD, Goorin AM, Epstein ML, Lipshultz SE. Clinical cardiotoxicity following anthracycline treatment for childhood cancer: the Pediatric Oncology Group experience. J Clin Oncol. 1997;15:1544–1552. doi: 10.1200/JCO.1997.15.4.1544. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien MM, Taub JW, Chang MN, Massey GV, Stine KC, Raimondi SC, Becton D, Ravindranath Y, Dahl GV Children's Oncology Group Study, P.O.G. Cardiomyopathy in children with Down syndrome treated for acute myeloid leukemia: a report from the Children's Oncology Group Study POG 9421. J Clin Oncol. 2008;26:414–420. doi: 10.1200/JCO.2007.13.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalabus JL, Sanborn CC, Jamil RG, Cheng Q, Blanco JG. Expression of the anthracycline-metabolizing enzyme carbonyl reductase 1 in hearts from donors with Down syndrome. Drug Metab Dispos. 2010;38:2096–2099. doi: 10.1124/dmd.110.035550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacological reviews. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 6.Olson RD, Mushlin PS, Brenner DE, Fleischer S, Cusack BJ, Chang BK, Boucek RJ., Jr Doxorubicin cardiotoxicity may be caused by its metabolite, doxorubicinol. Proc Natl Acad Sci U S A. 1988;85:3585–3589. doi: 10.1073/pnas.85.10.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minotti G, Recalcati S, Mordente A, Liberi G, Calafiore AM, Mancuso C, Preziosi P, Cairo G. The secondary alcohol metabolite of doxorubicin irreversibly inactivates aconitase/iron regulatory protein-1 in cytosolic fractions from human myocardium. Faseb J. 1998;12:541–552. doi: 10.1096/fasebj.12.7.541. [DOI] [PubMed] [Google Scholar]
- 8.Quinones-Lombrana A, Ferguson D, Hageman Blair R, Kalabus JL, Redzematovic A, Blanco JG. Interindividual Variability in the Cardiac Expression of Anthracycline Reductases in Donors With and Without Down Syndrome. Pharmaceutical research. 2014 doi: 10.1007/s11095-013-1267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov M, Kacevska M, Ingelman-Sundberg M. Epigenomics and interindividual differences in drug response. Clin Pharmacol Ther. 2012;92:727–736. doi: 10.1038/clpt.2012.152. [DOI] [PubMed] [Google Scholar]
- 10.Cubitt HE, Yeo KR, Howgate EM, Rostami-Hodjegan A, Barter ZE. Sources of interindividual variability in IVIVE of clearance: an investigation into the prediction of benzodiazepine clearance using a mechanistic population-based pharmacokinetic model. Xenobiotica. 2011;41:623–638. doi: 10.3109/00498254.2011.560294. [DOI] [PubMed] [Google Scholar]
- 11.Rostami-Hodjegan A, Tucker GT. Simulation and prediction of in vivo drug metabolism in human populations from in vitro data. Nat Rev Drug Discov. 2007;6:140–148. doi: 10.1038/nrd2173. [DOI] [PubMed] [Google Scholar]
- 12.Ahmed NK. Daunorubicin reductase activity in human normal lymphocytes, myeloblasts and leukemic cell lines. European journal of cancer & clinical oncology. 1985;21:1209–1213. doi: 10.1016/0277-5379(85)90017-3. [DOI] [PubMed] [Google Scholar]
- 13.Bogason A, Masquelier M, Lafolie P, Skogastierna C, Paul C, Gruber A, Vitols S. Daunorubicin metabolism in leukemic cells isolated from patients with acute myeloid leukemia. Drug metabolism letters. 2010;4:228–232. doi: 10.2174/187231210792928260. [DOI] [PubMed] [Google Scholar]
- 14.Barron-Vivanco BS, Rothenberg SJ, Medina-Diaz IM, Robledo-Marenco L, Rojas-Garcia AE, Hernandez-Cadena L, Poblete-Naredo I, Elizondo G, Albores A. AKRs expression in peripheral blood lymphocytes from smokers: the role of body mass index. Human & experimental toxicology. 2013;32:418–426. doi: 10.1177/0960327112455071. [DOI] [PubMed] [Google Scholar]
- 15.Lemieux N, Malfoy B, Forrest GL. Human carbonyl reductase (CBR) localized to band 21q22.1 by high-resolution fluorescence in situ hybridization displays gene dosage effects in trisomy 21 cells. Genomics. 1993;15:169–172. doi: 10.1006/geno.1993.1024. [DOI] [PubMed] [Google Scholar]
- 16.Hefti E, Quinones-Lombrana A, Redzematovic A, Hui J, Blanco JG. Analysis of mtDNA, miR-155 and BACH1 expression in hearts from donors with and without Down syndrome. Mitochondrial DNA. 2014:1–8. doi: 10.3109/19401736.2014.926477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Covarrubias V, Zhang J, Kalabus JL, Relling MV, Blanco JG. Pharmacogenetics of human carbonyl reductase 1 (CBR1) in livers from black and white donors. Drug Metab Dispos. 2009;37:400–407. doi: 10.1124/dmd.108.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical chemistry. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 19.Darzynkiewicz Z, Juan G. DNA content measurement for DNA ploidy and cell cycle analysis. Current protocols in cytometry / editorial board, J. Paul Robinson, managing editor … [et al.] 2001;Chapter 7(Unit 7):5. doi: 10.1002/0471142956.cy0705s00. [DOI] [PubMed] [Google Scholar]
- 20.P. McCullagh JAN. Generalized Linear Models. Chapman and Hall/CRC; 1989. [Google Scholar]
- 21.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic acids research. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic acids research. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 24.Kacevska M, Ivanov M, Wyss A, Kasela S, Milani L, Rane A, Ingelman-Sundberg M. DNA methylation dynamics in the hepatic CYP3A4 gene promoter. Biochimie. 2012;94:2338–2344. doi: 10.1016/j.biochi.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Adkins RM, Krushkal J, Tylavsky FA, Thomas F. Racial differences in gene-specific DNA methylation levels are present at birth. Birth defects research. Part A, Clinical and molecular teratology. 2011;91:728–736. doi: 10.1002/bdra.20770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trevor Hastie RT Jerome Friedman. The Elements of Statistical Learning. Springer-Verlag; 2009. [Google Scholar]
- 27.Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, Norrholm SD, Kilaru V, Smith AK, Myers AJ, Ramirez M, Engel A, Hammack SE, Toufexis D, Braas KM, Binder EB, May V. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu X, Li F, Yang B, Liang J, Qin H, Xu J. Effects of ultraviolet B exposure on DNA methylation in patients with systemic lupus erythematosus. Experimental and therapeutic medicine. 2013;5:1219–1225. doi: 10.3892/etm.2013.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baron B. Breaking the Silence: The Interplay Between Transcription Factors and DNA Methylation, in Methylation - From DNA, RNA and Histones to Diseases and Treatment. In: Dricu A, editor. INTECH, Biochemisty, Genetics, and Molecular Biology. 2012. p. 23. [Google Scholar]
- 30.Letourneau A, Santoni FA, Bonilla X, Sailani MR, Gonzalez D, Kind J, Chevalier C, Thurman R, Sandstrom RS, Hibaoui Y, Garieri M, Popadin K, Falconnet E, Gagnebin M, Gehrig C, Vannier A, Guipponi M, Farinelli L, Robyr D, Migliavacca E, Borel C, Deutsch S, Feki A, Stamatoyannopoulos JA, Herault Y, van Steensel B, Guigo R, Antonarakis SE. Domains of genome-wide gene expression dysregulation in Down's syndrome. Nature. 2014;508:345–350. doi: 10.1038/nature13200. [DOI] [PubMed] [Google Scholar]
- 31.Pope BD, Gilbert DM. Genetics: Up and down in Down's syndrome. Nature. 2014;508:323–324. doi: 10.1038/508323a. [DOI] [PubMed] [Google Scholar]
- 32.Heisey JMHaDM. The Abuse of Power: The Pervasive Fallacy of Power Calculations for Data Analysis. The American Statistician. 2001;55:19–24. [Google Scholar]
- 33.Grenier MA, Lipshultz SE. Epidemiology of anthracycline cardiotoxicity in children and adults. Seminars in oncology. 1998;25:72–85. [PubMed] [Google Scholar]
- 34.Lipshultz SE, Sambatakos P, Maguire M, Karnik R, Ross SW, Franco VI, Miller TL. Cardiotoxicity and cardioprotection in childhood cancer. Acta haematologica. 2014;132:391–399. doi: 10.1159/000360238. [DOI] [PubMed] [Google Scholar]
- 35.Trachtenberg BH, Landy DC, Franco VI, Henkel JM, Pearson EJ, Miller TL, Lipshultz SE. Anthracycline-associated cardiotoxicity in survivors of childhood cancer. Pediatric cardiology. 2011;32:342–353. doi: 10.1007/s00246-010-9878-3. [DOI] [PubMed] [Google Scholar]
- 36.Lavelle D, DeSimone J, Hankewych M, Kousnetzova T, Chen YH. Decitabine induces cell cycle arrest at the G1 phase via p21(WAF1) and the G2/M phase via the p38 MAP kinase pathway. Leukemia research. 2003;27:999–1007. doi: 10.1016/s0145-2126(03)00068-7. [DOI] [PubMed] [Google Scholar]
- 37.Tao J, Liu Q, Wu X, Xu X, Zhang Y, Wang Q, Luo C. Identification of hypermethylation in hepatocyte cell adhesion molecule gene promoter region in bladder carcinoma. Int J Med Sci. 2013;10:1860–1867. doi: 10.7150/ijms.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipshultz SE, Karnik R, Sambatakos P, Franco VI, Ross SW, Miller TL. Anthracycline-related cardiotoxicity in childhood cancer survivors. Current opinion in cardiology. 2014;29:103–112. doi: 10.1097/HCO.0000000000000034. [DOI] [PubMed] [Google Scholar]
- 39.Deng S, Wojnowski L. Genotyping the risk of anthracycline-induced cardiotoxicity. Cardiovascular toxicology. 2007;7:129–134. doi: 10.1007/s12012-007-0024-2. [DOI] [PubMed] [Google Scholar]
- 40.Visscher H, Ross CJ, Rassekh SR, Barhdadi A, Dube MP, Al-Saloos H, Sandor GS, Caron HN, van Dalen EC, Kremer LC, van der Pal HJ, Brown AM, Rogers PC, Phillips MS, Rieder MJ, Carleton BC, Hayden MR Canadian Pharmacogenomics Network for Drug Safety, C. Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J Clin Oncol. 2012;30:1422–1428. doi: 10.1200/JCO.2010.34.3467. [DOI] [PubMed] [Google Scholar]
- 41.Lubieniecka JM, Graham J, Heffner D, Mottus R, Reid R, Hogge D, Grigliatti TA, Riggs WK. A discovery study of daunorubicin induced cardiotoxicity in a sample of acute myeloid leukemia patients prioritizes P450 oxidoreductase polymorphisms as a potential risk factor. Frontiers in genetics. 2013;4:231. doi: 10.3389/fgene.2013.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Liu W, Sun CL, Armenian SH, Hakonarson H, Hageman L, Ding Y, Landier W, Blanco JG, Chen L, Quinones A, Ferguson D, Winick N, Ginsberg JP, Keller F, Neglia JP, Desai S, Sklar CA, Castellino SM, Cherrick I, Dreyer ZE, Hudson MM, Robison LL, Yasui Y, Relling MV, Bhatia S. Hyaluronan synthase 3 variant and anthracycline-related cardiomyopathy: a report from the children's oncology group. J Clin Oncol. 2014;32:647–653. doi: 10.1200/JCO.2013.50.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.