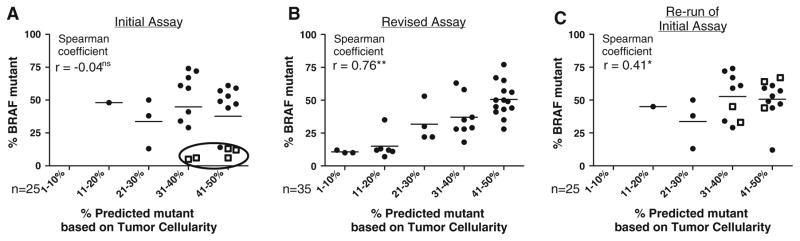
Fig. 3.
Correlations of predicted % mutant allele (based on tumor cellularity) and observed % mutant allele (detected by pyrosequencing). Initial pyrosequencing assay (a). Samples with low p.V600E BRAF mutations, despite high tumor cellularity (outliers) are circled. Squares represent v.600K-positive samples characterized as false, low positive p.V600E by the initial assay. Revised pyrosequencing assay (b): correlation for 35 p.V600E BRAF mutations. Repeat analysis of the 25 samples from the initial assay (c): re-run of the revised pyrosequencing assay. Significance *p < 0.05 or **p < 0.01