The α-synuclein gene, SNCA, is selectively expressed in blood cells and neurons. Locascio et al. reveal a paradoxical reduction in SNCA transcript counts in the blood of individuals with early-stage, neuroimaging-supported Parkinson’s disease in three regional, national, and international populations. Low SCNA transcript abundance predicted subsequent cognitive decline.
Keywords: α-synuclein, biomarker, gene expression, biobank, mitochondria, cognitive decline
The α-synuclein gene, SNCA, is selectively expressed in blood cells and neurons. Locascio et al. reveal a paradoxical reduction in SNCA transcript counts in the blood of individuals with early-stage, neuroimaging-supported Parkinson’s disease in three regional, national, and international populations. Low SCNA transcript abundance predicted subsequent cognitive decline.
Abstract
There are no cures for neurodegenerative diseases and this is partially due to the difficulty of monitoring pathogenic molecules in patients during life. The Parkinson’s disease gene α-synuclein (SNCA) is selectively expressed in blood cells and neurons. Here we show that SNCA transcripts in circulating blood cells are paradoxically reduced in early stage, untreated and dopamine transporter neuroimaging-supported Parkinson’s disease in three independent regional, national, and international populations representing 500 cases and 363 controls and on three analogue and digital platforms with P < 0.0001 in meta-analysis. Individuals with SNCA transcripts in the lowest quartile of counts had an odds ratio for Parkinson’s disease of 2.45 compared to individuals in the highest quartile. Disease-relevant transcript isoforms were low even near disease onset. Importantly, low SNCA transcript abundance predicted cognitive decline in patients with Parkinson’s disease during up to 5 years of longitudinal follow-up. This study reveals a consistent association of reduced SNCA transcripts in accessible peripheral blood and early-stage Parkinson’s disease in 863 participants and suggests a clinical role as potential predictor of cognitive decline. Moreover, the three independent biobank cohorts provide a generally useful platform for rapidly validating any biological marker of this common disease.
Introduction
Nearly 5 million people have Parkinson’s disease, a sporadic, progressive neurodegenerative disorder linked to a complex genetic architecture and environmental exposures. Over the next 15 years, this number will almost double to 9 million patients worldwide (Dorsey et al., 2007). Already today, the costs to public health are enormous—estimated at $10.8 billion annually in the USA alone (O'Brien et al., 2009). There are no cures or disease-modifying therapies for Parkinson’s disease, and this may be due in part to our inability to monitor biological markers of early disease. Currently, patients with Parkinson’s disease are diagnosed, cared for, and assigned to clinical trials based on medical history and physical exam.
To translate research progress into the clinic, simple, dynamic biomarkers are needed that are rooted in genetic discoveries. Traditionally, Parkinson’s disease has been approached from a clinical perspective, as a homogeneous movement disorder typified by the triad of resting tremor, bradykinesia and rigidity. Based on this clinical view of the disease, it was postulated that a single laboratory or imaging biomarker for Parkinson’s disease would be developed to serve as a universal diagnostic. This goal has been elusive. More recently, a human genome-inspired view of Parkinson’s disease has provided unequivocal evidence for a diversity of genetic variants, each exacting a small, but definitive effect, underlying the clinical phenotype (Nalls et al., 2014). It is becoming clearer that Parkinson’s disease is aetiologically, genetically, pathologically and even clinically more heterogeneous than previously implied (Scherzer and Feany, 2004; Scherzer et al., 2004; Forman et al., 2005). In light of this diversity the traditional expectation of a ‘one size fits all’ laboratory diagnostic that can detect all patients and answer all clinical questions may need to be revised. Personalized biomarkers, tailored to patient endophenotypes and specific clinical applications, are likely to emerge. They may more appropriately describe the molecular architecture of the disease.
What are the practical applications that such biomarkers may serve, if they cannot be used as a universal diagnostic? Prognostic and mechanism-directed biomarkers will help to stratify patients for clinical trials. Risk markers are required for identifying high-risk individuals prior to the onset of overt clinical symptoms, when disease-modifying interventions will be most effective. Pharmacodynamic markers are required to determine whether an investigational drug engages the intended target. In cancer drug development, biomarker use has increased average phase III trial success by 34% (Hayashi et al., 2013) and shortened total drug development timelines (Chin et al., 2011). Emerging CSF tests for Parkinson’s disease (Mollenhauer et al., 2011) and dopamine transporter imaging are relatively invasive or time-consuming and expensive. Blood-based tests are attractive, but no disease-causing molecule has been rigorously validated.
Accumulation of α-synuclein protein (encoded by the SNCA gene) in the brain is a hallmark of Parkinson's disease (Braak et al., 2002). For nearly two decades since its characterization as an electric ray protein, the Parkinson's disease gene α-synuclein was thought of as ‘neuron-specific’ (Maroteaux et al., 1988), but this view was disproven with the discovery of high levels of α-synuclein expression in blood cells, particularly those of erythroid lineage (Nakai et al., 2007; Scherzer et al., 2008; Maitta et al., 2011). Here we tested whether levels of SNCA transcripts (mRNAs) in circulating blood cells might serve to identify individuals with increased risk of Parkinson’s disease. We evaluated whether levels of α-synuclein gene (SNCA) transcripts are associated with the presence of early-stage, clinically and neuroradiologically confirmed Parkinson’s disease in a platform of three independent biomarker study cohorts.
Materials and methods
Brief methods are summarized below. See the online Supplementary material for detailed clinical, laboratory and statistical experimental methods for the Harvard Biomarker Study (HBS), Prognostic Biomarkers in Parkinson’s Disease Study (PROBE) and Parkinson’s Progression Markers Initiative (PPMI), respectively, and for the meta-analysis across three studies.
Biospecimen collections
Briefly, the studies were designed to minimize bias from sample processing by collecting, handling and analysing specimens of cases and controls in a standardized manner following defined operating procedures and according to rules of evidence (Ransohoff, 2004, 2005; Hennecke and Scherzer, 2008). Cases and controls were processed in parallel by technicians blinded to diagnosis to avoid bias due to ‘run order’ of samples. Venous whole blood was collected in PAXgene™ tubes. Blood and PAXgene™ reagents were mixed by gently inverting 8–10 times.
RNA isolation and quality control
RNA was extracted according to the PAXgene™ Blood RNA kit manual extraction protocol including DNase treatment. RNA quality was determined with the RNA 6000 Nano Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies) and the RNA Integrity Number package (Imbeaud et al., 2005).
Quantitative PCR and microarray expression analyses
Quantitative PCR was performed similar to Scherzer et al. (2008). Microarray procedures were performed as described (Zheng et al., 2010) using Illumina HumanHT-12v3 Expression BeadChips. The PROBE microarray data set has been submitted to the GEO database (accession number GSE57475).
NanoString assay performance
Probes were designed according to the manufacturer’s design principles (Geiss et al., 2008), including screening for inter- and intra-reporter and capture probe interactions, and selection for probes with optimal melting temperatures (Geiss et al., 2008). Direct counts of the target RNAs were measured in 125 ng of RNA by digital expression analysis based on NanoString technology (without reverse transcription into cDNA). Probes for the target and control RNAs were multiplexed and assayed on the nCounter Digital Analyzer. The laboratory running the assay was blinded to the diagnosis. No-template (negative) controls containing water substituted for template were run and no signal was detected. To avoid run-order bias, samples of cases or controls were randomly assigned to plates. To control for plate-to-plate variation and drift, equal amounts of Human Universal Reference RNA were included at the beginning, within, and at the end of the entire experiment. Transcript counts (excluding SNCA-007) measured in reference RNA were highly correlated with R2 > 0.999 both within one plate and in-between different plates, thus excluding drift as a potential source of bias in the experiment. Furthermore, 5% of participants’ samples were randomly resampled to verify the retest reliability (technical precision). The average R2 was 0.98 for these correlations. The NanoString data are accessible through the PPMI website (http://www.ppmi-info.org).
Results
Harvard Biomarker Study: SNCA mRNA abundance in early-stage Parkinson’s disease
We first evaluated relative SNCA mRNA abundance in a case-control study nested in the HBS using precise, kinetic, quantitative PCR based on fluorogenic 5’ nuclease chemistry (quantitative PCR). We specifically designed the HBS as a clinical biomarker study with rigorous, predefined collection and processing protocols. Cases and controls had similar ages, but patients with Parkinson’s disease were more likely to be males (Table 1). Cases were at an early stage of the disease, with a mean modified Hoehn and Yahr stage of 2.1. A large majority of cases (201 of 222, 90.5%) were on medications that ameliorate the dopamine deficiency caused by the degeneration of neurons in the substantia nigra, while 9.5% (21 of 222) were untreated, de novo patients. The controls were recruited from the same source population. Case and control samples were collected, processed, and analysed in parallel. Samples were required to meet stringent quality control criteria in order to enter the study including a RNA Integrity Number (RIN) (Auer et al., 2003) threshold of ≥7.3 indicating high RNA quality. The two groups had excellent RNA quality with mean RINs of 8 ± 0.4 versus 8.05 ± 0.4 [mean ± standard deviation (SD)], respectively.
Table 1.
The HBS: patient characteristics
| Patients with Parkinson’s disease | Controls | P-value | |
|---|---|---|---|
| (n = 222) | (n = 183) | ||
| Age at phlebotomy (years, SD) | 67.05 (9.53) | 68.44 (10.23) | 0.16 |
| Gender | |||
| Male (n, %) | 130 (58.6) | 74 (40.4) | 0.0003 |
| Female (n, %) | 92 (41.4) | 109 (59.6) | |
| Clinical findings (n, %) | |||
| UKPDBBSa | 207 (93.2) | ||
| Resting tremor | 164 (73.87) | ||
| Bradykinesia | 221 (99.59) | ||
| Rigidity | 219 (98.65) | ||
| Postural imbalance | 109 (49.10) | ||
| Asymmetric onset | 175 (78.83) | ||
| Medications (n, %) | |||
| Levodopa | 170 (76.58) | ||
| Dopamine replacement medications | 201 (90.5) | ||
| De novo | 21 (9.5%) | ||
| Disease severity | |||
| Disease duration (years) (mean, SD) | 4.8 (4.5) | ||
| Hoehn and Yahr (mean, SD) | 2.1 (0.6) | ||
| UPDRS Part I: mentation, behaviour, mood | 1.5 (1.5) | ||
| UPDRS Part II: ADLs | 8.7 (5.2) | ||
| UPDRS Part III: motor examination | 18.5 (9.0) | ||
| UPDRS total score | 30.9 (13.6) | ||
aUnited Kingdom Parkinson’s Disease Brain Bank Society diagnostic criteria.
ADL = activities of daily living.
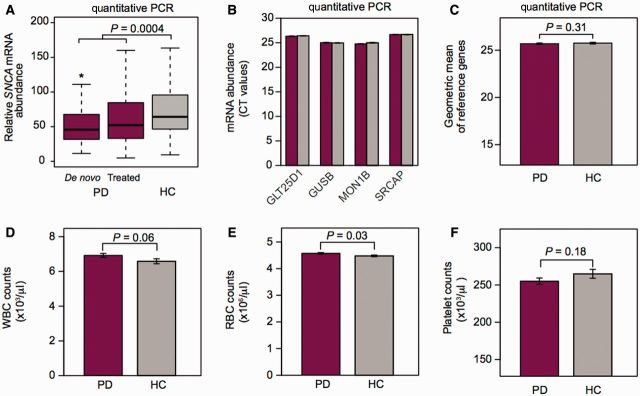
Relative SNCA transcript abundance in venous blood was significantly lower for patients with Parkinson’s disease than for controls (difference of 20%; P-value = 0.0004; Table 2 and Fig. 1A). Post hoc analysis of the subgroup of untreated, de novo cases showed consistent results with a >20% lower mean relative SNCA transcript abundance in the de novo cases compared to the controls (P-value = 0.01). Counts of the four endogenous reference genes (Fig. 1B) and their geometric mean (that was used to normalize for input RNA) were virtually identical in cases and controls (Fig. 1C). Importantly, complete blood counts in cases and controls could not account for the reduction in relative SNCA mRNA abundance in patients consistent with previous reports by others (Kasten et al., 2010). Specifically, white blood cell counts (Fig. 1D) and platelet counts (Fig. 1F) were unchanged between cases and controls, while red blood cell counts were marginally higher in cases than in controls (P = 0.03; Fig. 1E).
Table 2.
Blood SNCA expression is associated with early-stage clinical and DAT-neuroimaging supported Parkinson’s disease in three independent populations and on three independent assay platforms
| Cases | Controls | Difference of means | P-value* | |
|---|---|---|---|---|
| HBS | ||||
| Quantitative PCR platformc | ||||
| n (total n = 405) | 222 | 183 | ||
| Unadjusted relative SNCA expression (mean, SD) | 51.5 (1.97) | 64.2 (1.8) | −20e | 0.0004 |
| Adjusted relative SNCA expressiond (mean, 95% CI) | 49 (45–54) | 61 (55–68) | −17e | 0.003 |
| PROBE | ||||
| Quantitative PCR platformc | ||||
| n (total n = 118) | 76 | 42 | ||
| Unadjusted relative SNCA expression (mean, SD) | 75.6 (2.27) | 96.3 (1.7) | −22e | 0.025 |
| Adjusted relative SNCA expressiond (mean, 95% CI) | 72 (61–85) | 96 (76–122) | −25e | 0.046 |
| Microarray platform | ||||
| n (total n = 142) | 93 | 49 | ||
| ILMN-PROBE1a Unadjusted SNCA expression (mean, SD) | 2846 (1962) | 3469 (1622) | −18 | 0.01 |
| ILMN-PROBE1a Adjusted SNCA expressiond (mean, 95% CI) | 2452 (2167–2775) | 3098 (2635–3644) | −21 | 0.02 |
| ILMN-PROBE2b Unadjusted SNCA expression (mean, SD) | 681 (473) | 823 (432) | −17 | 0.02 |
| ILMN-PROBE2a Adjusted SNCA expressiond (mean, 95% CI) | 578 (508–657) | 730 (616–864) | −21 | 0.03 |
| PPMI | ||||
| Digital gene expression platform | ||||
| n (total n = 340) | 202 | 138 | ||
| Unadjusted analyses | ||||
| E3E4-SNCA transcript counts (mean, SD) | 13 458 (7812) | 15 959 (12 576) | −16 | 0.08 |
| E4E6-SNCA transcript counts (mean, SD) | 2826 (1860) | 3496 (3,119) | −19 | 0.04 |
| Long 3’UTR-SNCA transcript counts (mean, SD) | 573 (387) | 781 (1013) | −27 | 0.007 |
| SNCA-007 transcript counts (mean, SD) | 23 (16) | 25 (21) | −8 | 0.6 |
| Adjusted modelsd | ||||
| Adjusted E4E6-SNCA transcript counts (mean, 95% CI) | 2409 (2196–2643) | 2784 (2495–3107) | −13 | 0.046 |
| Adjusted long 3’UTR-SNCA transcript counts (mean, 95% CI) | 485 (440–534) | 578 (515–647) | −16 | 0.02 |
aILMN-PROBE1 indicates Illumina probe ILMN1701933 that targets the 3’UTR of SNCA.
bILMN-PROBE2 indicates Illumina probe ILMN1766165 that targets exon 5 of SNCA.
cAll statistical analyses for quantitative PCR assays were conducted using the natural logarithm of ΔCT values. For presentation purposes in Table 2 and Figs 1 and 2, a linearizing transformation of mean ΔCTs was performed. Values shown are in arbitrary units.
dAdjusted for covariates of gender, and white and red blood cell counts.
eIndicates per cent reduction in expression in cases compared to controls determined using the comparative CT method.
*Indicates significance analysis for all measurement values was performed on log-transformed data.
Figure 1.
HBS: reduced SNCA mRNA abundance in early-stage Parkinson’s disease. (A) Mean SNCA expression was significantly lower in 222 patients with early-stage clinical Parkinson’s disease (PD) compared to 183 controls (HC) without neurologic disease enrolled in the HBS using precise, quantitative PCR (unadjusted difference of 20%; P-value of 0.0004). *P-value = 0.01 for untreated, de novo cases versus controls. Box plots visualize first, third quartiles and means; the ends of the whiskers represent the lowest (or highest) value still within 1.5-times the interquartile range. (B) Expression values of the four endogenous reference genes used in the HBS were virtually identical in cases and controls. Cycle threshold (CT) values (standard error) are shown for cases (crimson bars) and controls (grey bars). (C) The geometric mean of these four endogenous reference genes was used to normalize for input RNA. (D–F) Importantly, complete blood counts in cases and controls could not account for the reduction in relative SNCA mRNA abundance in patients. Specifically, white blood cell counts (D, WBC) and platelet counts (F) did not significantly differ between cases and controls, while red blood cell counts (RBC) were marginally higher in cases than in controls (E; mean red blood cell, 4.6 versus 4.5, P = 0.03). Bar graphs show means and standard errors.
General linear model analysis was performed adjusting for the covariates of counts of white and red blood cells, and gender. In this covariate-adjusted analysis the mean relative abundance of SNCA expression was 17% lower in cases than in controls with P = 0.003.
Odds ratios (OR) for the lowest (first) quartile relative to the highest (fourth) quartile for blood SNCA transcript abundance were associated with Parkinson’s disease status in the unadjusted analysis and remained associated following adjustments for the covariates of age, hours at 4°C, red and white blood cells, and platelets (Model 1) or for gender (Model 2) with odds ratios ranging from 2.14 (95% CI, 1.10–4.14) to 2.15 (95% CI, 1.2–3.82) (Table 3).
Table 3.
The odds ratio for Parkinson’s disease prevalence is increased in individuals with blood SNCA expression in the lowest quartile of values compared to individuals with SNCA expression in the highest quartile of values in three study populations
| Quartile |
||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| HBS | ||||
| Relative SNCA mRNA abundance | 5–37a | 37–60a | 60–91a | 91–294a |
| Parkinson’s disease (n = 222), n/total (%) | 67/222 (30) | 58/222 (26) | 50/222 (23) | 47/222 (21) |
| Healthy control (n = 183), n/total (%) | 34/183 (18.6) | 43/183 (23.5) | 51/183 (27.9) | 55/183 (30) |
| Unadjusted OR, (95% CI)b | 2.31 (1.31–4.1) | 1.58 (0.91–2.75) | 1.15 (0.66–1.99) | 1 |
| P-value | 0.0036 | 0.11 | 0.63 | |
| Model 1, OR (95% CI)c | 2.14 (1.10–4.14) | 1.86 (1.01–3.42) | 1.07 (0.6–1.92) | 1 |
| P-value | 0.024 | 0.046 | 0.82 | |
| Model 2, OR (95% CI)d | 2.15 (1.2–3.82) | 1.61 (0.92–2.83) | 1.12 (0.64–1.96) | 1 |
| P-value | 0.0096 | 0.098 | 0.69 | |
| PROBEe | ||||
| Relative SNCA mRNA abundance | 16–52a | 56–79a | 84–119a | 119–2702a |
| Parkinson’s disease (n = 76), n /total (%) | 24/76 (32) | 20/76 (26) | 17/76 (22) | 15/76 (20) |
| Healthy control (n = 42), n/total (%) | 5/42 (12) | 10/42 (24) | 13/42 (31) | 14/42 (33) |
| OR (95% CI)b,f | 4.5 (1.3–15.0) | 1.9 (0.7–5.3) | 1.2 (0.4–3.4) | 1 |
| P-value | 0.02 | 0.90 | 0.25 | |
| PPMI | ||||
| Long-3’UTR-SNCA transcript counts | 59–328 | 335–497 | 505–786 | 796–10 861 |
| Parkinson’s disease (n = 202), n/total (%) | 58/202 (29) | 48/202 (24) | 55/202 (27) | 41/202 (20) |
| Healthy control (n = 138), n/total (%) | 27/138 (19.6) | 37/138 (26.8) | 30/138 (21.7) | 44/138 (31.9) |
| Unadjusted OR, 95% CI)b | 2.3 (1.2–4.3) | 1.4 (0.8–2.5) | 1.97 (1.1–3.6) | 1 |
| P-value | 0.009 | 0.28 | 0.03 | |
| Adjusted OR (95% CI)g | 2.1 (1.1–4.0) | 1.4 (0.8–2.6) | 2.1 (1.1–3.9) | 1 |
| P-value | 0.02 | 0.25 | 0.02 | |
aAll statistical analyses for quantitative PCR assays were conducted using ΔCT values. For presentation purposes the ranges of each quartile of values shown were linearly transformed, e.g. 2-lowest ΔCT and 2-highest ΔCT.
bUnadjusted odds ratio (OR) and confidence intervals (CI) were calculated using logistic regression models comparing the risk of each of quartiles 1 to 3 to the reference quartile 4 (highest relative SNCA transcript abundance).
cModel 1 consists of covariates age, hours in 4°C, red blood cells, platelets, and white blood cells in addition to the SNCA quartiles.
dModel 2 consists of covariate sex in addition to the SNCA quartiles.
ePROBE results on quantitative PCR platform are shown, similar results were obtained on the microarray platform.
fNo adjustments were indicated.
gModel consists of the covariate white blood cells in addition to the quartiles of long 3’UTR-SNCA counts.
PROBE study
Replication of low SNCA mRNA abundance in dopamine transporter imaging-supported Parkinson’s disease
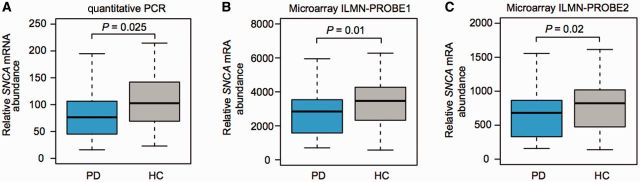
To be of use in clinical trials, it must be feasible to measure a biomarker in a multicentre study design. We thus further evaluated SNCA mRNA levels in the USA-wide, multicentre PROBE study (Supplementary Table 1). There were no significant baseline inequalities for age, gender, and white and red blood cell counts. Cases had a mean Hoehn and Yahr stage of 2.0, a mean total UPDRS score of 30.6, and mean disease duration of 5.5 years. The clinical diagnosis was supported by dopamine transporter (DAT) imaging with 123I-β-CIT. A subset of 76 cases with Parkinson’s disease and 42 controls with available high-quality RNA were analysed by quantitative PCR. The mean relative abundance of SNCA expression was 22% lower in cases than in controls with P = 0.025 in the unadjusted analysis (Table 2 and Fig. 2A). This remained significant after adjusting for the covariates of white and red blood cells, and gender with P = 0.046. Quantitative PCR allows for precise relative, but not absolute quantification. Thus, the difference in SNCA mRNA abundance in cases relative to controls found in PROBE was comparable to the 20% difference seen in HBS (i.e. in Figs 1A and 2A), while the absolute expression values (in arbitrary units) were not directly comparable due to different calibrators.
Figure 2.
USA-wide, multicentre PROBE study: replication of low SNCA mRNA abundance in DAT imaging-confirmed Parkinson’s disease. (A) We further evaluated this association in 76 cases with DAT imaging-confirmed Parkinson’s disease and 42 controls enrolled in the PROBE study. A 22% reduction in levels of blood-SNCA expression was seen in cases compared to controls on the quantitative PCR platform (P = 0.025). (B and C) These results were technically replicated on a second, independent microarray expression platform that includes two distinct probes for SNCA. Results for each of the two probes confirmed that relative blood-SNCA expression was lower in cases compared to controls. Box plots visualize first, third quartiles and means; the ends of the whiskers represent the lowest (or highest) value still within 1.5-times the interquartile range.
We confirmed these expression changes using two SNCA probes represented on a secondary, microarray expression platform. Results for both probes were available for 93 cases and 49 controls enrolled in PROBE and confirmed that relative blood SNCA expression was lower in cases compared to controls (Table 2 and Fig. 2B and C). Results for ILMN-PROBE1 confirmed an association between SNCA expression and Parkinson’s disease adjusting for the covariates of white and red blood cell counts, and gender with P = 0.02. The association was further confirmed by ILMN-PROBE2 with P = 0.03, again adjusting for the same covariates.
Individuals with SNCA transcript abundance in the lowest quartile of values had an OR for Parkinson’s disease of 4.5 (95% CI, 1.3–15.0) compared to individuals in the highest quartile of values (Table 3). The corresponding odds ratios for the two microarray probes were 4.4 (95% CI, 1.4–14.5) and 4.6 (95% CI, 1.5–13.7), respectively. Thus, measuring blood SNCA expression was feasible in a multicentre study and reduced blood SNCA expression in Parkinson’s disease was confirmed in this independent population on two distinct analysis platforms.
PPMI
Reduced abundance of disease-relevant SNCA isoforms already near disease onset
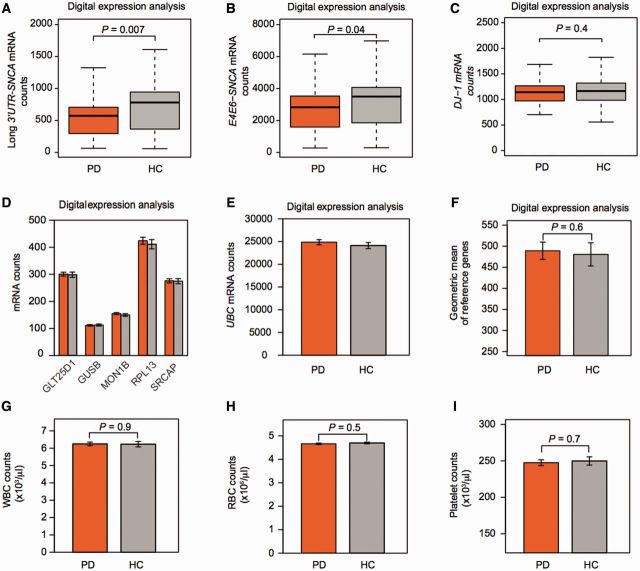
These associations prompted us to further extend and verify our findings in patients near disease onset, when individual motor symptoms are just beginning to manifest. Two hundred and two cases with de novo motor Parkinson’s disease, with less than 2 years of disease and with confirmed loss of dopaminergic substantia nigra terminals by DAT imaging and 138 age- and sex-matched controls without neurologic disease and with normal DAT imaging, who met stringent RNA quality-control criteria (RIN ≥ 7.3) were included. Key eligibility criteria for PPMI require the presence of asymmetric resting tremor or asymmetric bradykinesia, a supportive DAT scan, and a diagnosis of Parkinson’s disease for 2 years or less, without current dopamine replacement medication (Parkinson Progression Marker Initiative, 2011). Patients therefore were clinically at a very early stage and did not yet necessarily fulfil the standard, full clinical diagnostic criteria of the UK Parkinson’s Disease Society Brain Bank that require bradykinesia and a second mandatory clinical cardinal sign and three supportive clinical criteria to be present for diagnosis (Hughes et al., 1992). Cases and controls were frequency matched for age and gender (Supplementary Table 2). The PPMI population was remarkable for the virtual absence of Parkinson’s disease medications, and the equal distribution of potentially confounding variables of age, gender, and blood cell counts among cases and controls (Supplementary Table 2 and Fig. 3G–I).
Figure 3.
PPMI: disease-relevant SNCA isoforms are already reduced near disease onset. (A and B) SNCA isoforms were probed on a highly robust digital expression platform in 202 cases with de novo motor Parkinson’s disease (PD), with less than 2 years of disease and with confirmed loss of dopaminergic substantia nigra terminals by DAT imaging and 138 age- and sex-matched controls (HC) without neurological disease and normal DAT imaging participating in PPMI. Unadjusted counts of disease-relevant SNCA transcript isoforms with long 3' UTR (A) or skipping exon 5 (B), were reduced by 27% and 19%, respectively, in de novo cases compared to controls (P = 0.007 and 0.04, respectively). (C) By contrast mean counts for the gene PARK7 (DJ-1) mutated in rare autosomal recessive Parkinson’s disease were not significantly different in de novo cases with sporadic Parkinson’s disease compared to controls (difference of 1.5%). Box plots visualize first, third quartiles and means; the ends of the whiskers represent the lowest (or highest) value still within 1.5-times the interquartile range. (D–F) Reference genes were stably expressed in blood cells of cases compared to controls. Counts of the six endogenous reference genes used in PPMI (D and E) were virtually identical in the 202 cases and 138 controls. Mean counts (standard error) are shown for cases (orange bars) and controls (grey bars). (F) The geometric mean of these six endogenous reference genes was used to normalize for input RNA. It was nearly identical in cases and controls. (G–I) White (WBC) and red (RBC) blood cell, and platelet counts did not differ significantly between cases and controls. Bar graphs show means and standard errors.
The advent of digital expression analysis with molecular barcodes (Geiss et al., 2008; Malkov et al., 2009) allowed us to count the abundance of SNCA transcripts directly in RNA from blood cells, without the need for reverse transcription or PCR amplification. No signal was detected in controls lacking template. To avoid run-order bias, samples of cases or controls were randomly assigned to plates. Transcript counts (excluding SNCA-007) measured in reference RNA were highly correlated with R2 > 0.999 both within one plate and in-between different plates, thus excluding drift as a potential source of bias. Furthermore, 5% of participants’ samples were randomly resampled to verify the retest reliability (technical precision). The average R2 was 0.98 for these correlations.
Alternative SNCA transcript isoform usage has been suggested as a potential convergent mechanism in Parkinson’s disease pathology (Rhinn et al., 2012). We therefore designed multiple probes (Supplementary Fig. 1) to target the boundaries of exon 3 and exon 4 (henceforth termed E3E4-SNCA transcript isoform), transcripts specifically with a long 3’UTR region (which may redirect α-synuclein towards mitochondria; Rhinn et al., 2012), transcripts that skip exon 5 (termed E4E6-SNCA transcript isoform) and are translated into a truncated, aggregation-prone 112 amino acid protein (Murray et al., 2003), or the rare short SNCA-007 transcript isoform that comprises exons 1–4. Relative counts of long 3'UTR-SNCA transcripts were reduced by 27% in cases with Parkinson’s disease compared to controls in the unadjusted analysis with P = 0.007 (Table 2 and Fig. 3A). This was confirmed with a second digital molecular probe directed at a different target sequence in long-3’UTR-SNCA transcripts. The relation between long-3'UTR-SNCA transcripts and Parkinson’s disease remained significant with a P-value of 0.02 after adjusting for the covariates of white blood cell, red blood cell, and gender (Table 2). Counts of exon 5-skipping transcripts were also associated with Parkinson’s disease in unadjusted and adjusted analyses with P-values of 0.04 and 0.046, respectively (Table 2 and Fig. 3B). Counts for E3E4-SNCA transcripts were 16% lower in cases compared to controls but did not reach significance. Counts of the rare SNCA-007 transcript did not significantly differ between cases and controls. To estimate whether SNCA expression is specially associated with Parkinson’s disease or whether Parkinson’s disease-linked transcripts are generally perturbed in circulating blood cells of patients with Parkinson’s disease, we examined the expression of loci linked to familial Parkinson’s disease [PARK7 (also known as DJ-1) and PARK15 (also known as FBXO7)] and ZNF746 (implicated in mediating the effects of mutant PARK2; Shin et al., 2011). Furthermore, the QDPR gene produces an essential cofactor for dopamine biosynthesis and was also evaluated. These Parkinson’s disease-linked transcripts were similarly expressed in cases and controls with no or minimal fold-changes of <5% and non-significant P-values [although one of five PARK15/FBXO7 isoforms tested (FBXO7-001) showed a marginal trend with P = 0.06]. For example, mean counts for PARK7 mRNAs were 1144 in de novo cases and 1160 in controls (1.5% change, P = 0.4; Fig. 3C). Counts (Fig. 3D and E) and geometric mean (Fig. 3F) of the six endogenous reference genes (used to normalize for input RNA) were nearly identical in cases and controls. Cases and controls did not significantly differ in white cells, red cells, and platelets counts (Fig. 3G–I).
Individuals with long-3'UTR-SNCA transcript counts in the lowest quartile of values had an OR > 2 for Parkinson’s disease compared to individuals in the highest quartile of values in both the unadjusted analysis and analyses adjusted for white blood cell count, the only covariate retained in a step-wise backward elimination procedure (Table 3).
Risk of Parkinson’s disease associated with relative SNCA transcript abundance
Clinically useful biological markers of complex diseases can have modest areas under the curve (AUC) and a wide range of values that overlap in subjects with a clinical disease and in those without it (Manolio, 2003). Consistent with this notion, SNCA transcripts showed AUCs for Parkinson’s disease in the range of a possible risk marker. The AUC for SNCA transcripts as a predictor of Parkinson’s disease was 0.6 in HBS, ranged from 0.63 to 0.66 in PROBE (for quantitative PCR and microarray probes, respectively), and was 0.58 in PPMI.
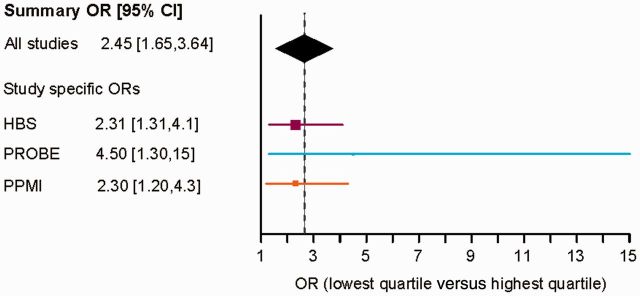
A meta-analysis was made possible by the similar collection protocols between the cohorts and allowed for powerfully combining the three studies to estimate the association between SNCA transcript abundance and risk of early-stage Parkinson’s disease. It increased the sample size to 500 cases and 363 controls (total of 863). SNCA variables estimated on quantitative PCR and digital expression platforms were standardized and ranked. The covariates of white and red blood cell count, gender, and age were adjusted for in the meta-analysis. For PPMI we first conservatively included expression measures for the generic probe (E3E4-SNCA) that targets most SNCA transcripts. In a second iteration, we included the specific probe targeting long 3'UTR-SNCA. In both scenarios the association between SNCA blood transcripts and early Parkinson’s disease was highly significant with P = 0.0001 and P < 0.0001, respectively. Overall, individuals with SNCA transcript levels in the lowest quartile had a highly significant OR for Parkinson’s disease of 2.18 (96% CI, 1.47–3.22) and 2.45 (96% CI, 1.65–3.64; Fig. 4) for each of the two scenarios, respectively. Cochran’s Q statistic equalled 2.67 (P = 0.26), giving no evidence of heterogeneity among the three studies.
Figure 4.
Risk of Parkinson’s is associated with relative SNCA transcript abundance. Individuals with SNCA transcript levels in the lowest quartile of values had a summary odds ratio for Parkinson’s disease of 2.45 (1.65, 3.64) compared to individuals with levels in the highest quartile in a meta-analysis across the three studies. In the Forrest plot squares represent the estimate of effect, i.e. the OR for each individual study, the size of the square corresponds to the size of the study, and bars around the square represent the 95% CI of the odds ratio. The diamond indicates the summary estimate; the width (the horizontal tips) of the diamond represents the 95% CI of the summary OR.
Taken together these results indicate that reduced SNCA mRNA levels in circulating cells are associated with Parkinson’s disease, even near disease onset and in patients with DAT-imaging supported loss of dopaminergic nigral projections. Exploration of specific transcript isoforms using digital transcript counting indicated that SNCA transcripts containing a long 3'UTR may be particularly relevant for this association.
SNCA transcript abundance predicts cognitive decline
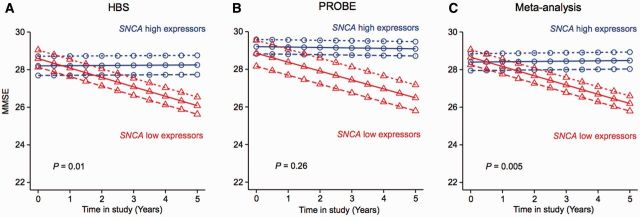
Predictors of cognitive decline would be valuable for stratifying clinical trials and for advancing clinical care (Chen-Plotkin, 2014). We have tracked Mini-Mental State Examination (MMSE) scores in the HBS and the PROBE populations during multiple longitudinal in-person follow-up visits for ∼5 years. To evaluate whether SNCA transcript levels measured at enrolment can longitudinally predict cognitive prognosis in Parkinson’s disease, we analysed the 222 patients with Parkinson’s disease that were longitudinally followed in the HBS. In HBS, MMSE scores were obtained during in-person study visits at enrolment, after 12, 24, and 60 months in the study. There was overall a significant relation between SNCA transcript abundance at enrolment and MMSE scores during the longitudinal follow-up period with P = 0.0007. We then tested whether low versus high SNCA expression status measured at enrolment could predict longitudinal cognitive decline in these patients. The 25% of patients with Parkinson’s disease with the highest SNCA transcript levels at enrolment (n = 55; termed henceforth ‘SNCA high expressors’) and the 25% of patients with the lowest SNCA transcript levels at enrolment (n = 55; termed ‘SNCA low expressors’) were compared (Fig. 5). Notably, at enrolment high and low SNCA expressors had nearly identical mean (SD) MMSE scores of 28.37 (1.95) versus 28.53 (2.28) (P = 0.3). Moreover, the two groups showed no statistically significant differences in other clinical characteristics at enrolment, including gender, age, disease duration, total Unified Parkinson’s Disease Rating Scale (UPDRS) score, Hoehn and Yahr stage, medications, and years of education (Supplementary Table 3). Notably, low SNCA expressors showed accelerated longitudinal cognitive decline compared to high expressors with P = 0.01 in the longitudinal mixed random and fixed effects model analysis (Locascio and Atri, 2011) adjusting for age, gender, disease duration upon enrolment, and years of education (Fig. 5A).
Figure 5.
SNCA transcript abundance predicts cognitive decline in patients with Parkinson’s disease during longitudinal follow-up. (A) The 25% patients with Parkinson’s disease with the highest SNCA transcript levels at enrolment into HBS (n = 55; ‘SNCA high expressors’) and the 25% of patients with the lowest SNCA transcript levels at enrolment in HBS (n = 55; ‘low SNCA expressors’) were compared. Illustrative mean scores on the MMSE across time predicted from the estimated fixed effect parameters in the mixed random and fixed effects model analysis (Locascio and Atri, 2011) are shown for SNCA high expressors and SNCA low expressors. Low SNCA expressors showed accelerated longitudinal cognitive decline compared to high expressors with P = 0.01 adjusting for age, gender, disease duration upon enrolment, and years of education. (B) Mixed effects model analysis for the 25% SNCA high expressors (n = 16) versus the 25% low expressors in PROBE (n = 16) adjusting for covariates indicated a similar, but non-significant trend. (C) In the combined analysis, SNCA expression status was robustly confirmed as a predictor of longitudinal cognitive decline in Parkinson’s disease with P = 0.005 after adjusting for pertinent covariates and study. Illustrative mean scores on the MMSE across time predicted by the fitted model in the longitudinal analysis for Parkinson’s disease patients in the lowest quartile of SNCA expression levels are shown as red triangles; values for Parkinson’s disease patients in the highest quartile of SNCA expression levels are represented as blue circles (solid lines indicate predicted values for subjects with a mean value of disease duration at enrolment; large-dashed lines indicate those for individuals with 1 SD longer disease duration at enrolment; and short-dashed lines indicate those for subjects with 1 SD shorter disease duration at enrolment).
We then asked whether low and high expressor status can also predict cognitive decline in PROBE. Prospective longitudinal clinical data were available for 65 of the 76 patients in PROBE with relative SNCA abundance measured by quantitative PCR at enrolment. Again, the 25% of patients with Parkinson’s disease with the highest SNCA transcript levels at enrolment into PROBE (n = 16 in PROBE) and the 25% of patients with the lowest SNCA transcript levels at enrolment (n = 16 in PROBE) were tested. High and low SNCA expressors had similar MMSE scores of 29.19 (0.98) versus 28.44 (2.78) at enrolment (P = 0.95) (Supplementary Table 2). Mixed effects analysis adjusting for the covariates of age, gender, disease duration upon enrolment, and years of education indicated a similar trend as observed in the larger HBS population, but this did not reach statistical significance, likely because of insufficient power (Fig. 5B). PPMI was not included in the longitudinal analysis because the multi-year longitudinal follow-up data needed are not yet available. Finally, to increase power, we performed a mixed random and fixed effects meta-analysis across the low and high expressors from each of the HBS and PROBE populations. The combined analysis robustly confirmed SNCA expression status as predictor of longitudinal cognitive decline in Parkinson’s disease with P = 0.005 after adjusting for pertinent covariates and for study (Fig. 5C). Similar analyses were performed to evaluate whether SNCA expression can also predict decline in longitudinal total UPDRS scores. These analyses were inconclusive and will require further evaluation in larger, longitudinally followed patient populations.
Discussion
We show here that SNCA transcript counts in circulating blood cells are reduced in early Parkinson’s disease—even in the landmark PPMI study that enrolled patients before they met standard clinical diagnostic criteria. The association between SNCA expression and Parkinson’s disease was generalizable across platforms and across populations (Table 2 and Figs 1–3). It was observed in studies from a single health care system (HBS), from 22 USA sites (PROBE), and from 24 USA and international sites (PPMI). This association remained significant when adjusted for confounding variables such as age, gender, white and red blood cell counts, Parkinson’s disease medications, or sample handling.
Biological markers of processes that lead to complex diseases such as Parkinson’s disease are not simply present or absent, but have a wide range of values that overlap in subjects with a clinical disease and in those without it (Manolio, 2003). For example, total cholesterol has modest specificity and sensitivity for predicting cardiovascular disease (with an area under the receiver operating curve of 0.59 in the Women’s Health Study; Ridker et al., 2000)—well short of the >90% specificity required for a bona fide diagnostic test. Nonetheless, biological markers of complex diseases can have powerful clinical applications. Cholesterol metabolism-related biomarkers have transformed cardiology. They enable early intervention with cholesterol-lowering medications because of the favourable risk/benefit ratio (low side-effect profile combined with clear benefits for reducing debilitating cardiovascular events). Levels of these commonly used biomarkers are also valuable in tracking therapeutic response and patient stratification. Thus, if confirmed in prospective studies, SNCA blood transcripts could be developed into future markers of risk, of patient stratification for clinical trials, and of response to novel therapeutics (particularly those targeting α-synuclein production or clearance).
We have begun to explore a clinical application for this biological marker as a predictor of cognitive decline. Cognitive decline is one of the most debilitating manifestations of disease progression in Parkinson’s and is closely correlated with the presence of α-synuclein protein-positive Lewy body burden throughout the neocortex (Braak et al., 2002). Predictors of cognitive decline would be valuable for stratifying clinical trials and for advancing clinical care (Chen-Plotkin, 2014). Several previous studies have examined the association between cognitive decline and biological markers in cross-sectional studies (Mollenhauer et al., 2014) or with a single longitudinal follow-up after 1–2 years (Siderowf et al., 2010; Chen-Plotkin et al., 2011). Longitudinal studies with repeated measurements provide more independent information than a single measurement (Hedeker and Gibbons, 2006; Locascio and Atri, 2011). In our study low SNCA transcript abundance at enrolment predicted cognitive decline in patients with Parkinson’s disease that were cognitively reassessed during multiple longitudinal visits over up to 5 years of follow-up with a P-value of 0.005 in the combined analysis of HBS and PROBE studies.
Beyond these translational implications, the association here observed offers an immediate clue towards unravelling the biological function of α-synuclein in health and disease. α-Synuclein had been thought of as a neuron-specific gene for decades, but this view was turned on its head with the discovery of high levels of α-synuclein present in blood cells, particularly in reticulocytes (circulating immature red blood cells) and during terminal erythroid differentiation and in megakaryocytes (Scherzer et al., 2008; Maitta et al., 2011), with low or no expression in other tissues (Maroteaux et al., 1988). In erythroid cells SNCA expression is regulated by the GATA-1 transcription factor in synchrony with machinery essential for incorporating iron into heme within mitochondria (Scherzer et al., 2008) thereby pointing at a biological link between three nodes of Parkinson’s disease: α-synuclein, mitochondria (Zheng et al., 2010), and iron (Rhodes and Ritz, 2008). Our study suggests that this link might be perturbed in early disease stages. Alternative α-synuclein transcript isoform usage has been suggested as a potential convergent mechanism in Parkinson’s disease pathology (Rhinn et al., 2012). 3'UTR ends of mRNAs are critical for targeting transcripts to specific subcellular compartments and for translational control (Andreassi and Riccio, 2009). Specific transcript isoforms of SNCA with an extended 3'UTR impact accumulation of α-synuclein protein, which appears redirected away from synaptic terminals and towards mitochondria (Rhinn et al., 2012). This might be relevant to disease processes as mitochondrial dysfunction is implicated in the onset of Parkinson’s disease neuropathology (Zheng et al., 2010). By contrast, deletion of exon 5 of the SNCA gene results in the translation of a truncated 112 amino acid protein that is prone to aggregation and increases seeding efficiency (Murray et al., 2003).
In sporadic Parkinson’s disease, α-synuclein protein (and iron) accumulates in the brain and peripherally α-synuclein aggregates are found in skin, glands, intestines and epicardium (reviewed in Jellinger, 2011). In a few dozen patients a triplication of the SNCA locus causes monogenic familial Parkinson’s disease. The direction of the change observed here was therefore surprising, even paradoxical, at first glance. How can it be that SNCA blood transcript levels are consistently reduced in each of the three studies? Ample precedent exists for paradoxical changes in peripheral markers of neurological diseases. In Parkinson’s disease, extracellular α-synuclein protein levels are reduced in CSF for reasons unknown (Mollenhauer et al., 2011). In Wilson’s disease, copper accumulates in the basal ganglia, but serum levels are paradoxically low. In Alzheimer’s disease, amyloid-β42 levels are reduced in CSF (Hansson et al., 2006). We hypothesize that intracellular SNCA transcript levels in clinically manifest, sporadic Parkinson’s disease may be reduced as a consequence of intracellular α-synuclein protein accumulation (due to impaired clearance or other mechanisms) that leads to a feedback repression of transcription—in a homeostatic effort to ameliorate the ‘log jam’ of excessive α-synuclein protein products. In an alternate view, because SNCA is so highly expressed in reticulocytes, reduced SNCA expression could be a marker of generally abnormal reticulocyte biogenesis or function in early-stage Parkinson’s disease. We performed association studies that do not imply causality and that were not designed to distinguish between these alternative hypotheses. However, neither ours nor previous studies (Kasten et al., 2010) found evidence for an association between anaemia and Parkinson’s disease. By contrast, anaemia during early life, preceding disease onset by up to 20–29 years (Savica et al., 2009), and multiple recent blood donations have been correlated to increased risk of Parkinson’s disease (Logroscino et al., 2006).
The ease and non-invasiveness of an α-synuclein blood test is appealing. Previous attempts to search for α-synuclein blood tests centred on α-synuclein plasma protein, involved small cohorts, and were inconclusive (for review see Chahine et al., 2014). That approach poses considerable technical challenges as plasma can be easily contaminated with α-synuclein released from erythrocytes ruptured during sample transport and centrifugation. Our studies leapfrogged this technical challenge. Patient blood was directly collected into an FDA-approved system for preserving ‘intracellular RNA from whole blood for RT-PCR used in molecular diagnostic testing’ (without a need for plasma separation) where chemicals immediately stabilize the in vivo gene transcription profile by reducing in vitro RNA degradation and minimizing gene induction (Pahl and Brune, 2002a, b; Rainen et al., 2002; Kim et al., 2014). The HBS, PROBE and PPMI studies were purposefully designed to minimize the threats of bias from sample collection, processing and storage (Ransohoff, 2004). Case and control samples for HBS and PPMI were collected, processed and analysed in parallel using standardized procedures for each step. In contrast to other studies, RNA integrity, complete blood counts, and several processing and quality indicators were carefully measured for all three studies.
Although each of the three biomarker studies here presented has limitations when taken on its own, these individual constraints are compensated for by the collective strengths. For example, while the PROBE and HBS study could not completely rule out an undue influence of Parkinson’s disease medications on SNCA expression, this question was fully addressed by exclusively recruiting de novo cases in PPMI. We clearly show that this is not simply a marker of sick patients; instead, the association is already detectable in individuals who are very early in the disease course and highly functional. Clinical diagnostic certainty was enhanced through DAT neuroimaging.
Beyond revealing an association between lowered SNCA expression and Parkinson’s disease, this study presents a generally useful resource of three purpose-designed biobanks and linked clinical data. This platform of biobanks can now be used for accelerating the exploration of nearly any biofluid marker for Parkinson’s disease by decreasing the time from discovery to replication from multiple years down to the assay run-time.
Supplementary Material
Acknowledgements
We thank all study participants, their families, and friends for their support and participation, and our study coordinators.
Harvard Biomarker Study. Co-Directors: Harvard NeuroDiscovery Center: Clemens R. Scherzer, Bradley T. Hyman, Adrian J. Ivinson; Investigators and Study Coordinators: Harvard NeuroDiscovery Center: Ana Trisini-Lipsanopoulos, Kaltra Dhima, Stephen Bayer, Kaitlin C. Lockhart; Brigham and Women’s Hospital: Lewis R.Sudarsky, Michael T. Hayes, Reisa Sperling; Massachusetts General Hospital: John H. Growdon, Michael A. Schwarzschild, Albert Y. Hung, Alice W. Flaherty, Deborah Blacker, Anne-Marie Wills, U. Shivraj Sohur, Vivek K. Unni, Nicte I. Mejia, Anand Viswanathan, Stephen N. Gomperts, Vikram Khurana, Mark W. Albers, Kyleen E. Swords, Rebecca K. Rudel; University of Ottawa: Michael G. Schlossmacher; Scientific Advisory Board: Massachusetts General Hospital: John H. Growdon, Brigham and Women’s Hospital: Dennis J. Selkoe, Reisa Sperling; Harvard School of Public Health: Alberto Ascherio; Data Coordination: Harvard NeuroDiscovery Center: Thomas Yi, Massachusetts General Hospital: Joseph J. Locascio; Biobank Management Staff: Harvard NeuroDiscovery Center: Karen Duong, Zhixiang Liao, Ashley N. Hoesing.
PROBE Study. PROBE Steering Committee: Voyager Therapeutics: Bernard Ravina; Brigham and Women’s Hospital: Clemens Scherzer, University of Ottawa: Michael Schlossmacher, Avid Radiopharmaceuticals: Andrew Siderowf, University of Rochester: David Oakes; Institute for Neurodegenerative Disorders: Kenneth Marek; Georgetown University: Ira Shoulson. The investigators who contributed to the PROBE cohort and collection of clinical data and blood samples are listed in the Appendix of Ravina et al., A Longitudinal Program for Biomarker Development in Parkinson’s Disease: A Feasibility Study, Movement Disorders, Vol. 24, No. 14, 2009, pp. 2081–2090.
PPMI Study. The PPMI data used in this study were obtained from the Parkinson’s Progression Markers Initiative (PPMI) database (www.ppmi-info.org/data). For up-to-date information on the study, visit www.ppmi-info.org.
Glossary
Abbreviations
- DAT
dopamine transporter
- HBS
Harvard Biomarker Study
- MMSE
Mini-Mental State Examination
- PPMI
Parkinson’s Progression Markers Initiative
- PROBE
Prognostic Biomarkers in Parkinson’s Disease Study
Funding
This study was made possible by NIH grants U01 NS082157 (C.R.S.), U01 NS082080 (C.R.S.) the Harvard NeuroDiscovery Center (to A.I., C.R.S., and B.T.H.), U.S. Department of Defense grants W81XWH-1-0007 (B.R.) and W81XWH-13-1-0115 (C.R.S), and the Michael J. Fox Foundation (C.R.S. and M.G.S.). Additional funding was provided by the M.E.M.O. Hoffman Foundation (C.R.S.) and U.S. Department of Defense grant W81XWH-11-1-0150 (M.A.S.).
The Harvard Biomarker Study is supported by the Harvard NeuroDiscovery Center (HNDC), the Parkinson’s Disease Biomarkers Program (PDBP) grant U01 NS082157 of the NINDS, and the Massachusetts Alzheimer’s Disease Research Center (ADRC) P50 AG005134 grant of the National Institute on Aging.
The PROBE cohort was funded by Department of Defense Neurotoxin Exposure Treatment Parkinson’s Research Program (W23RRYX7022N606), NINDS PD-DOC (NS050095), the Parkinson’s Disease Foundation (New York, NY), with initial funding from Cephalon Inc and Lundbeck for the parent PRECEPT clinical trial and follow-up PostCEPT cohort, from which the nested PROBE cohort was derived.
PPMI, a public-private partnership, is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, including Abbott, Avid, Biogen Idec, Bristol-Myers Squibb, Covance, Elan, GE Healthcare, Genentech, GlaxoSmithKline, Lilly, Merck, Meso Scale Discovery, Pfizer, Roche, UCB.
The funders had no role in design or conduct of the study, collection, management, analysis and interpretation of the data, and preparation, review, or approval of the manuscript. The PPMI Data and Publications Review Committee reviewed the manuscript.
Supplementary material
Supplementary material is available at Brain online.
References
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol 2009; 19: 465–74. [DOI] [PubMed] [Google Scholar]
- Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Kornacker K. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat Genet 2003; 35: 292–3. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Bratzke H, Hamm-Clement J, Sandmann-Keil D, Rub U. Staging of the intracerebral inclusion body pathology associated with idiopathic Parkinson's disease (preclinical and clinical stages). J Neurol 2002; 249 (Suppl 3): III/1–5. [DOI] [PubMed] [Google Scholar]
- Chahine LM, Stern MB, Chen-Plotkin A. Blood-based biomarkers for Parkinson's disease. Parkinsonism Relat Disord 2014; 20 (Suppl 1): S99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS. Unbiased approaches to biomarker discovery in neurodegenerative diseases. Neuron 2014; 84: 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Hu WT, Siderowf A, Weintraub D, Goldmann Gross R, Hurtig HI, et al. Plasma epidermal growth factor levels predict cognitive decline in Parkinson disease. Ann Neurol 2011; 69: 655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med 2011; 17: 297–303. [DOI] [PubMed] [Google Scholar]
- Dorsey ER, Constantinescu R, Thompson JP, Biglan KM, Holloway RG, Kieburtz K, et al. Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology 2007; 68: 384–6. [DOI] [PubMed] [Google Scholar]
- Forman MS, Lee VM, Trojanowski JQ. Nosology of Parkinson's disease: looking for the way out of a quagmire. Neuron 2005; 47: 479–82. [DOI] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008; 26: 317–25. [DOI] [PubMed] [Google Scholar]
- Hansson O, Zetterberg H, Buchhave P, Londos E, Blennow K, Minthon L. Association between CSF biomarkers and incipient Alzheimer's disease in patients with mild cognitive impairment: a follow-up study. Lancet Neurol 2006; 5: 228–34. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Masuda S, Kimura H. Impact of biomarker usage on oncology drug development. J Clin Pharm Ther 2013; 38: 62–7. [DOI] [PubMed] [Google Scholar]
- Hedeker D, Gibbons RD. Longitudinal data analysis. Hoboken, NJ: Wiley; 2006. [Google Scholar]
- Hennecke G, Scherzer CR. RNA biomarkers of Parkinson’s disease: developing tools for novel therapies. Biomark Med 2008; 2: 41–53. [DOI] [PubMed] [Google Scholar]
- Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry 1992; 55: 181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, et al. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res 2005; 33: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinger KA. Synuclein deposition and non-motor symptoms in Parkinson disease. J Neurol Sci 2011; 310: 107–11. [DOI] [PubMed] [Google Scholar]
- Kasten M, Tadic V, Klein C, Rocca WA, Savica R, Eric Ahlskog J, et al. Anemia or low hemoglobin levels preceding Parkinson disease: a case-control study. Neurology 2010; 74: 1655; author reply 1656. [DOI] [PubMed] [Google Scholar]
- Kim JH, Jin HO, Park JA, Chang YH, Hong YJ, Lee JK. Comparison of three different kits for extraction of high-quality RNA from frozen blood. Springerplus 2014; 3: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio JJ, Atri A. An overview of longitudinal data analysis methods for neurological research. Dement Geriatr Cogn Dis Extra 2011; 1: 330–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logroscino G, Chen H, Wing A, Ascherio A. Blood donations, iron stores, and risk of Parkinson's disease. Mov Disord 2006; 21: 835–8. [DOI] [PubMed] [Google Scholar]
- Maitta RW, Wolgast LR, Wang Q, Zhang H, Bhattacharyya P, Gong JZ, et al. Alpha- and beta-synucleins are new diagnostic tools for acute erythroid leukemia and acute megakaryoblastic leukemia. Am J Hematol 2011; 86: 230–4. [DOI] [PubMed] [Google Scholar]
- Malkov VA, Serikawa KA, Balantac N, Watters J, Geiss G, Mashadi-Hossein A, et al. Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res Notes 2009; 2: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. Novel risk markers and clinical practice. N Engl J Med 2003; 349: 1587–9. [DOI] [PubMed] [Google Scholar]
- Maroteaux L, Campanelli JT, Scheller RH. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci 1988; 8: 2804–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer B, Locascio JJ, Schulz-Schaeffer W, Sixel-Doring F, Trenkwalder C, Schlossmacher MG. alpha-Synuclein and tau concentrations in cerebrospinal fluid of patients presenting with parkinsonism: a cohort study. Lancet Neurol 2011; 10: 230–40. [DOI] [PubMed] [Google Scholar]
- Mollenhauer B, Rochester L, Chen-Plotkin A, Brooks D. What can biomarkers tell us about cognition in Parkinson's disease? Mov Disord 2014; 29: 622–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IV, Giasson BI, Quinn SM, Koppaka V, Axelsen PH, Ischiropoulos H, et al. Role of alpha-synuclein carboxy-terminus on fibril formation in vitro. Biochemistry 2003; 42: 8530–40. [DOI] [PubMed] [Google Scholar]
- Nakai M, Fujita M, Waragai M, Sugama S, Wei J, Akatsu H, et al. Expression of alpha-synuclein, a presynaptic protein implicated in Parkinson's disease, in erythropoietic lineage. Biochem Biophys Res Commun 2007; 358: 104–10. [DOI] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet 2014; 46: 989–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien JA, Ward A, Michels SL, Tzivelekis S, Brandt NJ. Economic burden associated with Parkinson disease. Drug Benefit Trends 2009; 21: 179–90. [Google Scholar]
- Pahl A, Brune K. Gene expression changes in blood after phlebotomy: implications for gene expression profiling. Blood 2002a; 100: 1094–5. [DOI] [PubMed] [Google Scholar]
- Pahl A, Brune K. Stabilization of gene expression profiles in blood after phlebotomy. Clin Chem 2002b; 48: 2251–3. [PubMed] [Google Scholar]
- Parkinson Progression Marker Initiative. The Parkinson Progression Marker Initiative (PPMI). Prog Neurobiol 2011; 95: 629–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, et al. Stabilization of mRNA expression in whole blood samples. Clin Chem 2002; 48: 1883–90. [PubMed] [Google Scholar]
- Ransohoff DF. Rules of evidence for cancer molecular-marker discovery and validation. Nat Rev Cancer 2004; 4: 309–14. [DOI] [PubMed] [Google Scholar]
- Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer 2005; 5: 142–9. [DOI] [PubMed] [Google Scholar]
- Rhinn H, Qiang L, Yamashita T, Rhee D, Zolin A, Vanti W, et al. Alternative alpha-synuclein transcript usage as a convergent mechanism in Parkinson's disease pathology. Nat Commun 2012; 3: 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes SL, Ritz B. Genetics of iron regulation and the possible role of iron in Parkinson's disease. Neurobiol Dis 2008; 32: 183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker PM, Hennekens CH, Buring JE, Rifai N. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N Engl J Med 2000; 342: 836–43. [DOI] [PubMed] [Google Scholar]
- Savica R, Grossardt BR, Carlin JM, Icen M, Bower JH, Ahlskog JE, et al. Anemia or low hemoglobin levels preceding Parkinson disease: a case-control study. Neurology 2009; 73: 1381–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer CR, Feany MB. Yeast genetics targets lipids in Parkinson's disease. Trends Genet 2004; 20: 273–7. [DOI] [PubMed] [Google Scholar]
- Scherzer CR, Grass JA, Liao Z, Pepivani I, Zheng B, Eklund AC, et al. GATA transcription factors directly regulate the Parkinson's disease-linked gene alpha-synuclein. Proc Natl Acad Sci USA 2008; 105: 10907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer CR, Jensen RV, Gullans SR. Simplifying complex neurodegenerative diseases by gene chip analysis. In: Freese A, Simeone FA, Leone P, Janson C, editors. Principles of molecular neurosurgery. Basel: Karger; 2004. p. 246–57. [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, et al. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell 2011; 144: 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siderowf A, Xie SX, Hurtig H, Weintraub D, Duda J, Chen-Plotkin A, et al. CSF amyloid {beta} 1-42 predicts cognitive decline in Parkinson disease. Neurology 2010; 75: 1055–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson's disease. Sci Transl Med 2010; 2: 52ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.