Figure 3.
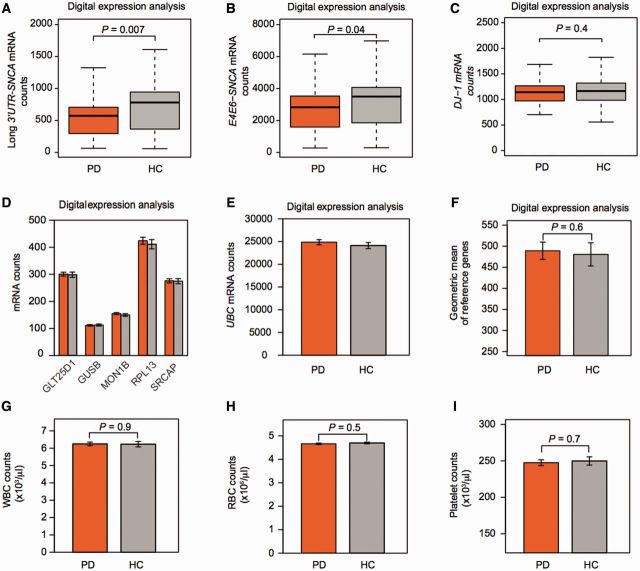
PPMI: disease-relevant SNCA isoforms are already reduced near disease onset. (A and B) SNCA isoforms were probed on a highly robust digital expression platform in 202 cases with de novo motor Parkinson’s disease (PD), with less than 2 years of disease and with confirmed loss of dopaminergic substantia nigra terminals by DAT imaging and 138 age- and sex-matched controls (HC) without neurological disease and normal DAT imaging participating in PPMI. Unadjusted counts of disease-relevant SNCA transcript isoforms with long 3' UTR (A) or skipping exon 5 (B), were reduced by 27% and 19%, respectively, in de novo cases compared to controls (P = 0.007 and 0.04, respectively). (C) By contrast mean counts for the gene PARK7 (DJ-1) mutated in rare autosomal recessive Parkinson’s disease were not significantly different in de novo cases with sporadic Parkinson’s disease compared to controls (difference of 1.5%). Box plots visualize first, third quartiles and means; the ends of the whiskers represent the lowest (or highest) value still within 1.5-times the interquartile range. (D–F) Reference genes were stably expressed in blood cells of cases compared to controls. Counts of the six endogenous reference genes used in PPMI (D and E) were virtually identical in the 202 cases and 138 controls. Mean counts (standard error) are shown for cases (orange bars) and controls (grey bars). (F) The geometric mean of these six endogenous reference genes was used to normalize for input RNA. It was nearly identical in cases and controls. (G–I) White (WBC) and red (RBC) blood cell, and platelet counts did not differ significantly between cases and controls. Bar graphs show means and standard errors.