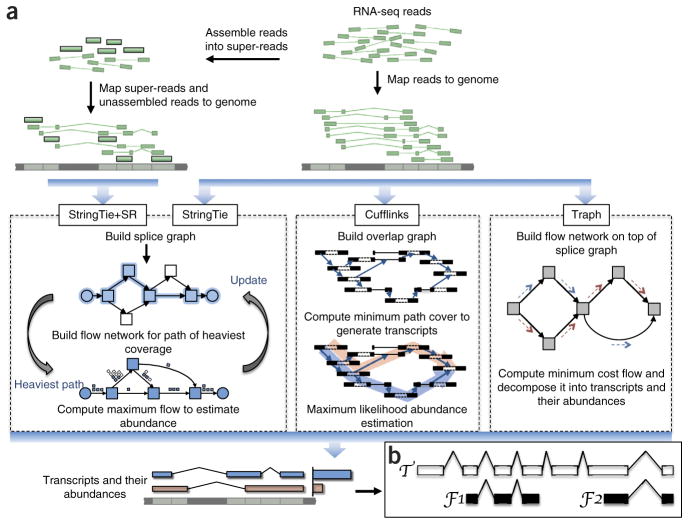
Figure 1.
Transcript assembly pipelines for StringTie, Cufflinks and Traph. (a) Overview of the flow of the StringTie algorithm, compared to Cufflinks and Traph. All methods begin with a set of RNA-seq reads that have been mapped to the genome. An optional secondary input to StringTie is a set of pre-assembled super-reads, designated as StringTie+SR. StringTie iteratively extracts the heaviest path from a splice graph, constructs a flow network, computes maximum flow to estimate abundance, and then updates the splice graph by removing reads that were assigned by the flow algorithm. This process repeats until all reads have been assigned. (b) Annotated transcript T for which read data covers only the fragments F1 and F2. An assembler is given credit for a correct reconstruction of T if it correctly assembles F1 and F2.