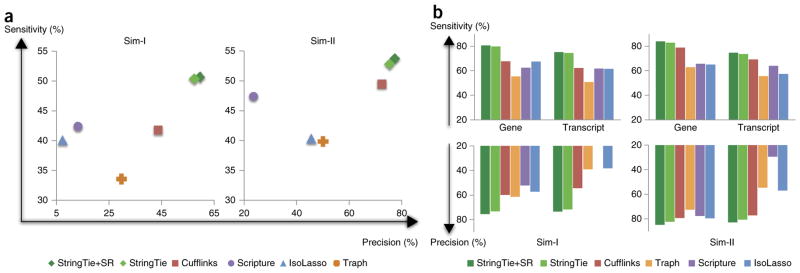
Figure 2.
Transcriptome assemblers’ accuracies in detecting expressed transcripts from two simulated RNA-seq data sets. (a) Transcriptome assemblers’ accuracies in detecting expressed transcripts from two simulated RNA-seq data sets. In data set Sim-I (left), the fragment sizes follow an empirical distribution based on Illumina sequences, and in Sim-II (right) the fragment sizes follow a parameterized normal distribution. StringTie+SR pre-assembles the reads into super-reads when possible. (b) Accuracy of transcriptome assemblers on gene loci from the same two data sets, considering only those transcripts that were completely covered by input reads. Scripture’s precision on Sim-I was 17.7%, below the 20% minimum shown here.