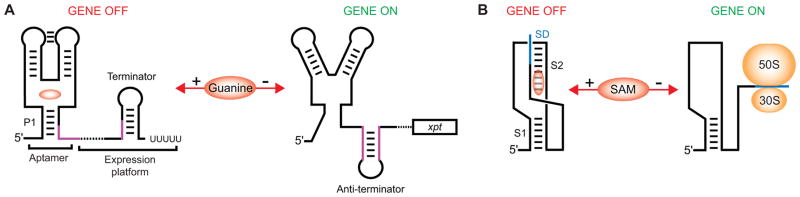
Fig. 1.
Typical mechanisms of the riboswitch-mediated gene expression control. (A), Transcription termination mechanism of the guanine-specific xpt riboswitch [73]. Guanine binding stabilizes the structure of the junctional metabolite-sensing domain, including regulatory helix P1, and facilitates the formation of a Rho-independent transcriptional terminator that aborts transcription elongation. In the absence of guanine, the metabolite sensor does not adopt the ligand-bound conformation, and the 3′ region of P1 (magenta color) makes alternative base pairing with the region in the expression platform inducing formation of an anti-terminator hairpin. This hairpin prevents formation of the transcription terminator and allows RNA polymerase to transcribe the entire xpt gene. (B), Regulation of translation initiation by the SAM-II riboswitch [52, 53]. SAM binding stabilizes the formation of the pseudoknot-based fold including stem S2 that engages in base pairing with the Shine-Dalgarno sequence (blue color) of the downstream gene. In the absence of SAM, S2 is disrupted and the ribosome can access the Shine-Dalgarno sequence and initiate translation of the gene.