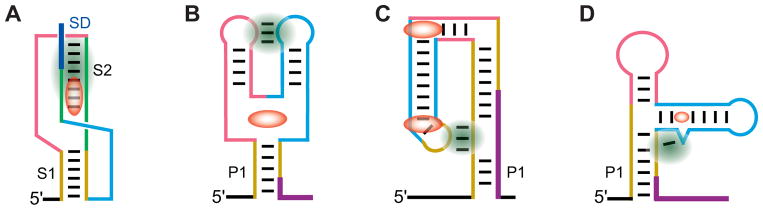
Fig. 2.
Schematics of architectures observed in riboswitch structures. Ligands are shown in red color. Long-distance tertiary interactions reinforced by ligand binding are highlighted in green color. (A), The pseudoknot type of the metabolite sensing domain, exemplified by the SAM-II riboswitch [52]. (B), The ‘regular’ architecture of junctional riboswitches observed in purine riboswitches. The fold is stabilized by ligand binding in the center of the three-way junction. (C), The ‘inverted’ architecture of junctional riboswitches stabilized in the THF riboswitch by binding to two ligand molecules [89]. (D), Glycine riboswitch is an example of the ‘regular’ architecture stabilized by tertiary interactions that span shorter distance [87].