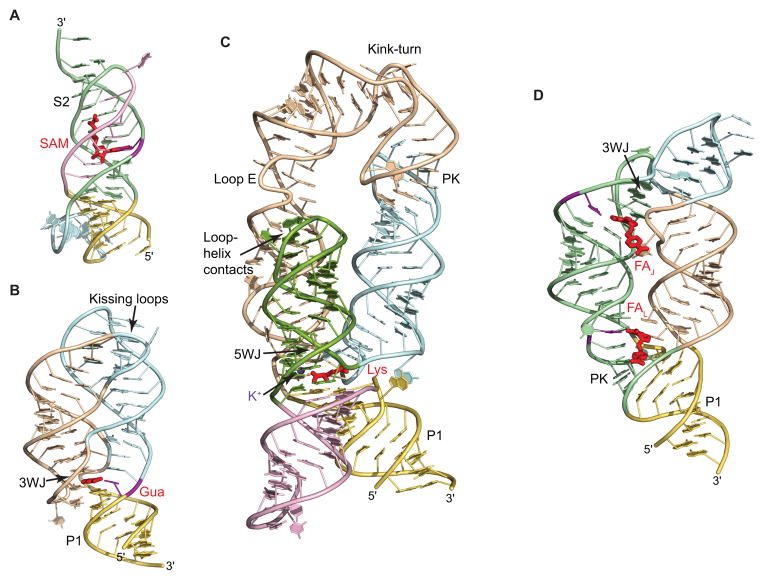
Fig. 3.
Examples of the riboswitch structures. RNA is shown in ribbon representation and is color-coded according to Fig. 2. Ligands are shown in red color. Nucleotides important for discrimination of the ligand are in violet. 3WJ and 5WJ, three-way and five-way junctions, respectively. PK, pseudoknot. (A) Structure of the SAM-II riboswitch bound to SAM highlights a riboswitch fold based on a pseudoknot conformation [52]. A key feature of the structure is the stabilization of stem S2 by binding to SAM. (B) Structure of the guanine riboswitch bound to guanine (Gua) shows ‘regular’ architecture of junctional riboswitches [73]. The ligand binds in the center of the three-way junction and holds together the junctional fold. Tertiary interactions between apical loops provide correct alignment of stems and reinforce the structure. (C) Structure of the lysine riboswitch bound to lysine [79]. The structure consists of two-helical and three-helical bundles positioned below and above the ligand-bound five-way junction. One of the helices reverses its orientation through a kink-turn and is anchored in place by pseudoknot interactions. Ligand-bound potassium cation is shown by a violet sphere. (D), Structure of the THF riboswitch bound to two molecules of folinic acid (FA) highlights the ‘inverted’ architecture of junctional riboswitches, centered on the three-way junction and stabilized by pseudoknot interactions [89]. Ligand molecules stabilize both the junction and the pseudoknot.