Figure 2. Abundance Changes in Specific Components of the Proteostasis Network.
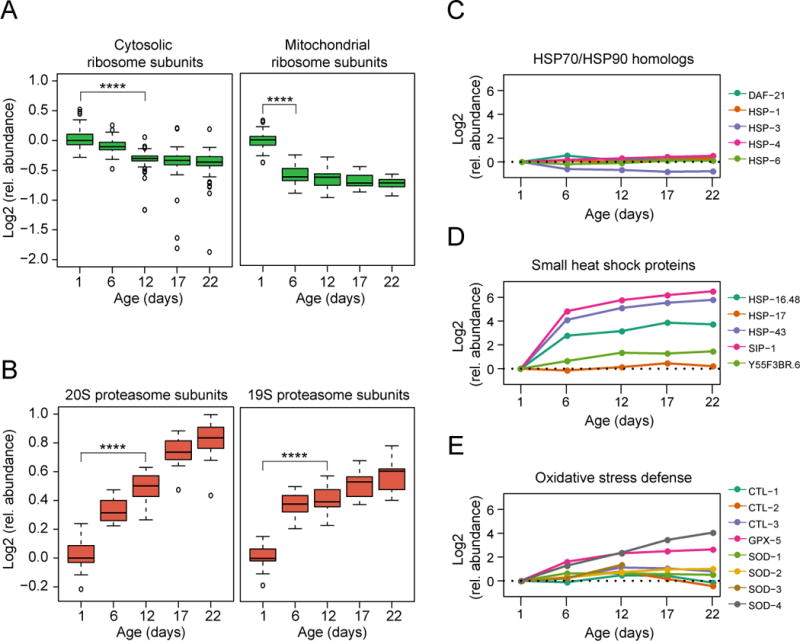
(A) Abundance changes of ribosomal proteins during the lifespan of C. elegans. 70 different cytosolic (left) and 34 mitochondrial ribosomal proteins (right) were quantified (see Table S3). Log2 values of fold-changes are shown in boxplot representation. Solid horizontal lines indicate the median values, whisker caps indicate 10th and 90th percentiles, and circles indicate outliers. ****, p-value <4.35 × 10−13 for cytosolic ribosomal proteins and 1.17 × 10−10 for mitochondrial ribosomal proteins from Wilcoxon signed rank test. Only proteins quantified at both time points tested were considered.
(B) Abundance changes of proteasome subunits during lifespan. All 14 subunits of the 20S and 17 subunits of the 19S proteasome were quantified. Only subunits quantified in at least two time points are displayed. ****, p-value <1.23 × 10−4 and ****, p-value <1.53 × 10−5 from Wilcoxon signed rank test.
(C–E) Abundance profiles of proteostasis network (PN) components along the lifespan of WT animals. Log2 relative changes in abundance are shown for HSP70 and HSP90 homologs (C), small HSPs (D), and proteins involved in oxidative stress defense (E). Only components quantified at day 1 and at least three consecutive time points are displayed.
