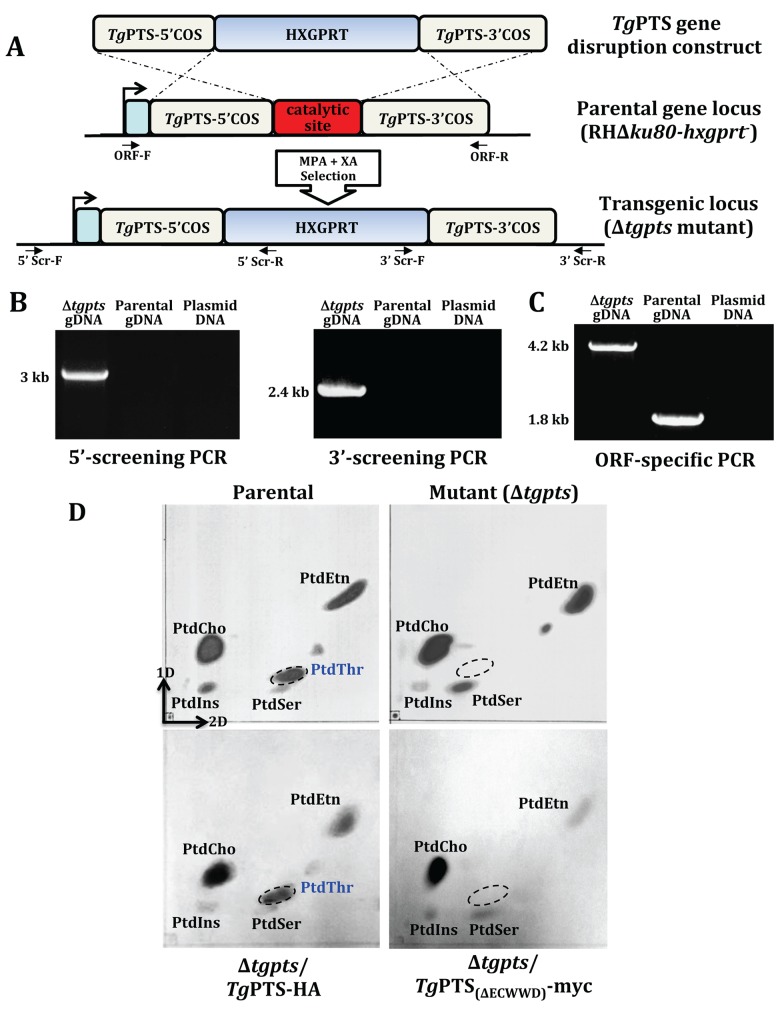
Fig 3. The Δtgpts strain is devoid of de novo PtdThr synthesis.
(A) Scheme for the targeted disruption of the TgPTS gene by double homologous recombination. The plasmid contained 5’ and 3’ crossover sequences (COS) flanking the hypoxanthine-xanthine-guanine phosphoribosyltransferase (HXGPRT) marker, which allows resistance to mycophenolic acid (MPA) and xanthine (XA). The PTS-disrupted (Δtgpts) strain lacking the conserved ECWWD (Glu-Cys-Trp-Trp-Asp) residues was identified by 5’ and 3’ screening primers (5’Scr-F/R, 3’Scr-F/R). (B) PCR images of a typical Δtgpts mutant showing specific amplification of the DNA bands by 5’ (3 kb) and 3’ (2.4 kb) genomic screening. The parental gDNA and plasmid were included as negative controls. (C) ORF-specific PCR confirming a successful insertion of the selection marker at the TgPTS gene locus. PCR shows amplification of an expected 4.2 kb band in the Δtgpts strain as opposed to the expected 1.8 kb in the parental parasites, and none in the plasmid DNA (negative control). Identity of all PCR amplicons was confirmed by sequencing. (D) Lipid profiles of the indicated strains by two-dimensional TLC. Total lipids (0.8–1 x 108 parasites) were resolved and detected by iodine-vapor staining. PtdThr band (encircled) is absent in the Δtgpts strain. It is restored by a wild-type TgPTS (TgPTS-HA), but not by a catalytically-inactive ΔECWWD isoform (TgPTS(ΔECWWD)-myc). Note that it appears as though PtdSer is not increased too much in the TgPTS(ΔECWWD)-myc-complemented mutant when compared to the parental strain. A sustained culture of the Δtgpts/TgPTS(ΔECWWD)-myc strain has proven particularly difficult due to severe growth defect (S9 Fig), likely caused by a dominant-negative effect exerted on PSS in an already attenuated mutant.