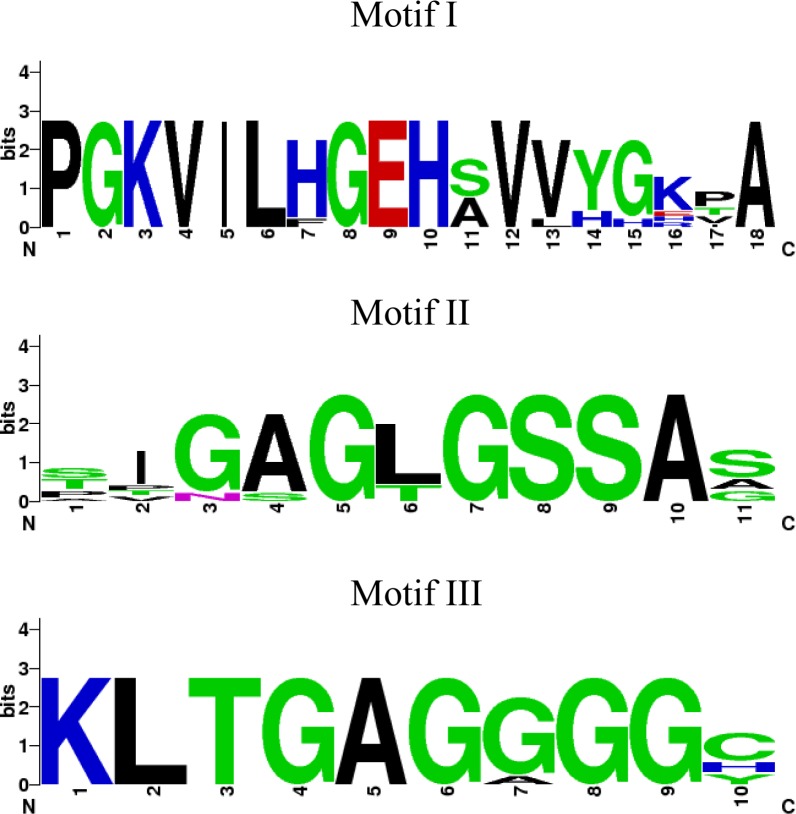
Fig 1. Motifs derived from MVK amino acid sequence alignments display consensus sequences.
Sequence logos for motif I, II and II were built using Weblogo [27]. The overall height of the stack indicates the sequence conservation at that position, colors denote the chemical properties of the amino acids. The sequences used are: Aedes aegypti (AAEL006435), Culex quinquefasciatus (EDS42994), Anopheles gambiae (EAA14782), Drosophila melanogaster (AGB93455), Bombyx mori (NP_001093299), Danaus plexippus (EHJ79258), Apis mellifera (XP_006558673), Acyrthosiphon pisum (XP_001942835) and Rattus norvegicus (NP_112325). Colors denote the chemical properties of the amino acids shown in each motif. Polar: G, S, T, Y, C; Neutral: Q; Basic: K, R, H; Acid: D, E; Hydrophobic: A, V, L, I, P, W, F, M.