Figure 1.
FANCD2 Protects Cells from Replication Stress and Preserves Genomic Integrity
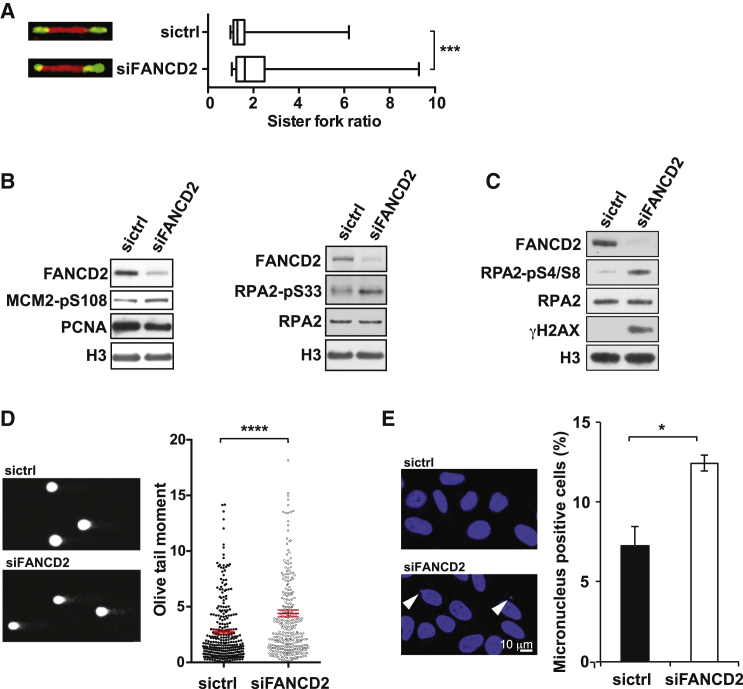
(A) DNA fiber analysis comparing sister fork symmetry. Shown are typical sister forks of U2OS cells treated with control (sictrl) or FANCD2 siRNA (siFANCD2). The ratios of the lengths of two corresponding sister replication forks are plotted. The middle line represents the median and the boxes the 25th and 75th percentiles. The whiskers mark the smallest and largest values. Mann-Whitney test was used to determine statistical significance (n = 3). ∗∗∗p ≤ 0.001.
(B) Western blots of whole-cell lysates of control and FANCD2 siRNA-treated U2OS cells probed for phosphorylation of MCM2 on Ser-108 (MCM2-pS108) and RPA2 on Ser-33 (RPA2-pS33). Histone H3, PCNA, and RPA2 served as loading controls.
(C) Western blot showing activation of the DDR upon depletion of FANCD2, including phosphorylation of histone H2AX on Ser-139 (γH2AX) and of RPA2 on Ser-4 and Ser-8 (RPA2-pS4/S8). RPA2 and histone H3 were used as loading controls.
(D) Comet assays of RNAi-treated U2OS cells. Individual data points of olive tail moment are plotted, showing mean ±SEM in red (n = 3, two-tailed Mann-Whitney test). ∗∗∗∗p ≤ 0.0001.
(E) DAPI-stained nuclei and micronuclei (arrowheads) of RNAi-treated U2OS cells. Mean ±SEM of micronucleus-positive cells are plotted (n = 3; unpaired, two-tailed Student’s t test). ∗p ≤ 0.05.