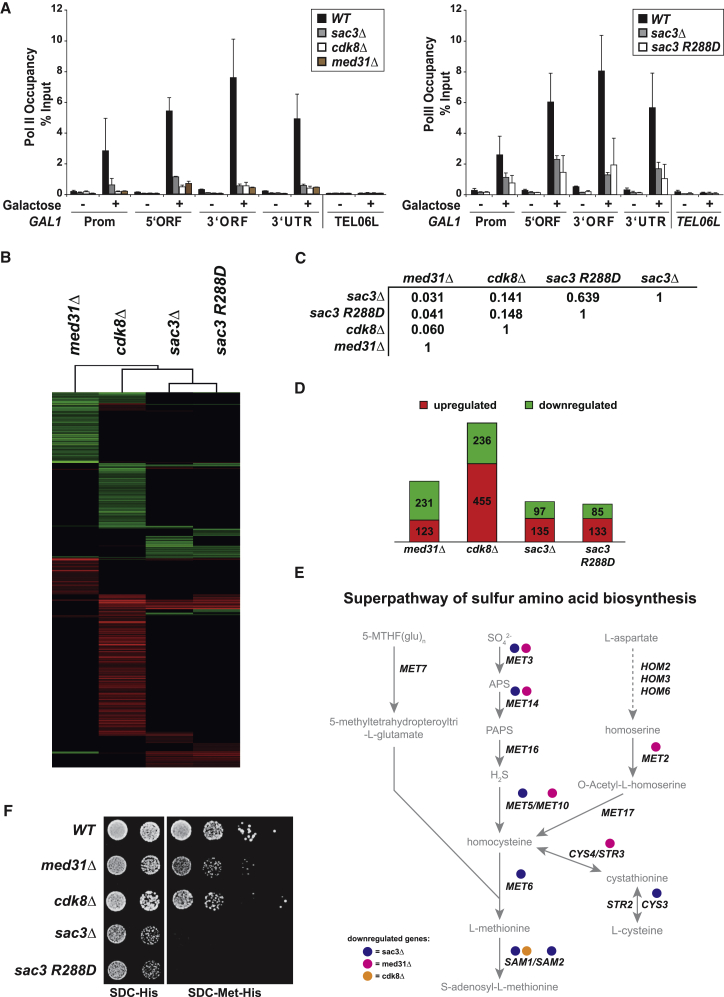
Figure 5.
The Sac3 PCI Domain Is Required for Transcription of Specific Genes In Vivo
(A) Pol II occupancy at the GAL1 promoter (Prom), 5′ORF, 3′ORF, and 3′UTR region was analyzed by ChIP in WT and mutant cells. Cells were either grown in raffinose or induced with 2% galactose for 120 min. Pol II occupancy at telomere 06L is shown as a negative control. Error bars represent SD of three independent experiments. qPCR primers are listed in Table S2.
(B) Cluster diagram of genes with significantly altered mRNA levels (>2.0-fold) in Mediator or TREX-2 mutant cells. Changes in mRNA levels compared to the wild-type strain are depicted in red (up), green (down), or black (no change).
(C) Pearson’s correlation matrix for expression profiles of TREX-2 and Mediator mutant strains as indicated (FC > 2.0 and FDR < 0.05).
(D) Number of significantly altered genes of all investigated strains. See also Figure S3C.
(E) Schematic view of sulfur amino acid biosynthesis superpathway. Genes with reduced expression in sac3Δ, med31Δ, or cdk8Δ cells are indicated.
(F) Growth analysis of WT, cdk8Δ, med31Δ, and sac3 mutant strains on medium with and without methionine. All BY4741 strains were transformed with a pRS411 (MET) plasmid to allow growth on methionine-deficient media.
See also Figures S4 and S7A and Table S5.