Figure S4.
ChIP-exo of TREX-2, Related to Figure 5
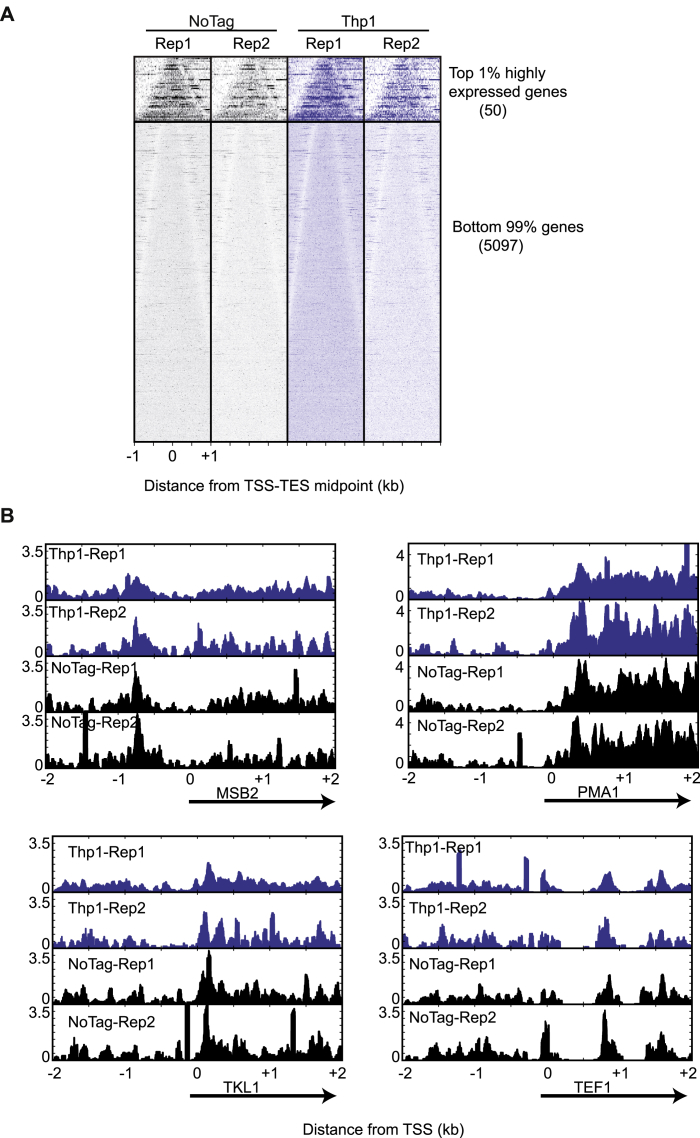
(A) Comparison of ChIP-exo profiles for Thp1-TAP and NoTag control in two independent experiments showed no significant difference in Thp1 occupancy. Bell plots show 5′-end tags for Thp1 and noTag control, aligned by the midpoint of transcription start site (TSS) and transcription end site (TES) and sorted by the gene length. Upper and lower sets of panel correspond to top 1% highly expressed genes and the remaining 99% genes. Bell plots are a graphical way to analyze if a particular factor is enriched at the 5′ or 3′ end of genes or throughout the gene body. For every transcript defined by (Xu et al., 2009), the 5′ end of the sequencing reads (tags) were retrieved for a defined region (+/− 2kb) around the midpoint of transcription start site (TSS) and transcription end site (TES). After plotting the tags with respect to the TSS-TES midpoint, the genes are sorted based on transcript length which gives it a characteristic bell shape. Both NoTag and Thp1-TAP show enrichment in the gene body relative to the 5′/3′ end of genes.
(B) Plots of Thp1 and NoTag ChIP-exo reads over putative TREX-2 target genes featured in (Santos-Pereira et al., 2014). Shown is the smoothed distribution of ChIP-exo tags 5′ ends over a 4kb window around the TSS of MSB2, PMA1, TKL1, and TEF1. All datasets were normalized to have an equal number of tags.